Distribution of human gene polymorphisms allele frequencies associated with viral infections
- Authors: Vlasenko N.V.1, Chanyshev M.D.2, Dubodelov D.V.1, Serkov A.A.1, Solopova G.G.3, Sacuk A.V.3, Snicar A.V.4, Semenenko T.А.5, Kuzin S.N.1, Akimkin V.G.1
-
Affiliations:
- Central Research Institute for Epidemiology, Federal Service for Surveillance on Consumer Rights Protection and Human Wellbeing
- Central Research Institute of Epidemiology, Federal Service for Surveillance on Consumer Rights Protection and Human Wellbeing
- Dmitry Rogachev National Medical Research Center of Pediatric Hematology
- Demikhov Moscow City Clinical Hospital
- N.F. Gamaleya National Research Center of Epidemiology and Microbiology
- Issue: Vol 68, No 5 (2023)
- Pages: 404-414
- Section: ORIGINAL RESEARCHES
- URL: https://virusjour.crie.ru/jour/article/view/14732
- DOI: https://doi.org/10.36233/0507-4088-189
- EDN: https://elibrary.ru/bguoyy
- ID: 14732
Cite item
Abstract
Introduction. The design of studies aimed at finding the association between the genetic factor and the studied feature (disease) involves a comparison of the ratio of genotypes or allelic proportions in the study group with those in the control group. At the stage of determining the ratio of genotypes of the studied polymorphisms in the reference group, researchers meet a number of problems, which are the subject of the present work.
Aim of the work is to provide scientific rationale for the feasibility of creating a national information system comprising genetic data of the relatively healthy population of Russia, incorporating its ethnic diversity.
Materials and methods. The study group, total 1020 people, was genotyped for a number of single nucleotide polymorphisms of human genes. A comparative characteristic of the frequency distribution of the studied polymorphisms with those presented in international databases as reference data was carried out using χ2 index.
Results. The frequency of SNP rs4986790 of the TLR4 gene significantly differs from the EUR population (p = 0.032) and the CEU subpopulation (p = 0.047). The allele frequencies of the rs1800795 (IL6) and rs1800896 (IL10) polymorphisms in the study population differ from the CEU subgroup (p = 0.030 and 0.012, respectively). The frequency of SNP rs2295119 (HLA-DPA2) in the study group is significantly different from the EUR population (p = 0.034).
Conclusion. The analysis carried out in this work confirms the need to create a domestic information system containing data on the occurrence of SNP alleles and genotypes for a conditionally healthy population and in subgroups with various pathological conditions.
Keywords
Full Text
Introduction
Free movement of people and increasing intermixing of populations are seen as distinctive features of the modern world and, in the opinion of experts, they contribute to reduction in the genetic diversity within the entire human population. Dunn et al. [1] believe that if the current trend remains unchanged for 2000 years (approximately 75 generations), it can result in a genetically uniform population of the planet. In the meantime, the genetic diversity in the world population is currently so high that epidemiological studies cannot be conducted ignoring the racial, ethnic, and even subethnic affiliation of the target population. Today, the study of historical, evolutionary, and present-day aspects of human development as well as human interaction with the natural environment is a priority avenue of world science. An increasing attention is given to the search of genetic determinants that play a critical role in selecting a personified approach when providing medical care to patients with various pathologies, especially with autoimmune and infectious diseases. The polyetiology of many human diseases, the complexity of pathogenic mechanisms, and a wide variety of forms (from acute forms with recovery and asymptomatic cases to fulminant forms and severe chronic types with malignant forms, etc.), the unpredictability of outcomes, and differences in susceptibility to infectious agents have shaped the conceptual view of the impact of genetic factors on development of pathological processes.
The RF Government Decree No. 715, 1/12/2004, On Approval of the List of Socially Significant Diseases and the List of Diseases Posing a Threat to Other People, highlighted significant diseases of viral etiology: tuberculosis, virus hepatitis B and C, HIV, and sexually transmitted infections. Addressing the course and outcomes of diseases caused by these infectious agents, studies on detection of specific molecular and genetic markers that are universal for autoimmune and infectious diseases demonstrate the significance of the problem.
The design of studies in assessment of the relationship of any genetic factor with the studied symptom (disease) unfailingly involves comparative analysis of the genotype frequencies or allele proportions in target and control groups. The main requirement for the control group is its full compliance with the ethnic structure of the population in the area where the study is performed. As a rule, the control group comprises individuals representing indicator groups of a relatively healthy population (blood donors, etc.) having no studied symptom (disease). The relevance criteria for the selected group are based on the assessment of the distribution of allele and genotype frequencies among the relatively healthy population against the existing data recognized as a benchmark.
Based on the statistics for 2022, the total population of Russia is 146.9 million people. The population is characterized by extremely high ethnic diversity represented by 180 nationalities living in the country. Such high historical-geographical and socio-cultural diversity of the population cannot produce the uniform distribution of genotypes within the population, which is confirmed by many works on ethnogenetics, one of the originators of which is Russian researcher Yu. Rychkov. In the monograph “The Gene Pool of the Population of Russia and Neighboring Countries” published under his editorship, close attention is given to some protein systems of the human body and their polymorphism contributing to significant differences in frequencies of the identified genotypes among population in different regions of Russia [2].
The high ethnic diversity of Russia poses a challenge to researchers in measuring proportions of genotypes of the studied polymorphisms in the comparison group, thus highlighting the need for an information system in Russia, which would contain data on frequencies of human gene polymorphisms in different regions of the country.
The purpose of the study is to provide scientific rationale for the feasibility of creating a national information system comprising genetic data of the relatively healthy population of Russia, incorporating its ethnic diversity.
Materials and methods
The study was performed at the viral hepatitis laboratory of the Central Research Institute of Epidemiology of the Federal Service for Consumer Rights Protection and Human Welfare (Rospotrebnadzor). The study group composed of relatively healthy citizens of Moscow was formed to estimate frequencies of alleles of some single nucleotide polymorphisms (SNPs). Based on the selection criteria, the enrollees were aged over 18 and had no infectious diseases. The study group included healthcare workers from the Treatment and Rehabilitation Center of the Ministry of Health of Russia (n = 283), blood donors of the Dmitry Rogachev National Medical Research Center of Pediatric Hematology, Oncology and Immunology (n = 427), healthcare workers from the Demikhov Municipal Clinical Hospital (n = 310). Since the size of a sample group increases its quantitative representativeness, the total number of participants was 1,020 people [3]. The study group was formed from Moscow healthcare workers and blood donors with Slavic participants accounting for the largest percentage. Inflows of migrants generally affect other sectors such as construction, municipal services, etc. However, in the healthcare sector, migrants are characterized by low numbers; therefore, in terms of the ethnic composition, the deviation in the sample subsets was minimum and did not produce any significant effect on the results. All the participants signed their informed consent for participation in the study. All samples were tested using the enzyme immunoassay for markers of infectious diseases including socially significant diseases such as virus hepatitis B and C, as well as hepatitis A and E, measles, polio, and chickenpox. DNA was extracted from peripheral blood using a Gemolitik reagent and a Ribo-PREP reagent kit (AmpliSens, Russia). The following SNP-containing genes were selected for the study: TLR4 (rs4986790), MERTK (rs4374383), IL1B (rs1143634, rs1143627), IL1RN (rs4251961, rs419598), IFNL4 (rs12979860, rs8099917), IL6 (rs1800795), IL10 (rs1800896), SERPINA1 (rs28929474), HLADPA2 (rs2295119), TULP1 (rs9380516). The above genes encode interleukins (ILs) involved in cascades of biochemical reactions associated with the inflammatory process occurring in various pathological conditions as well as polymorphisms, which are actively studied for their role in diseases of infectious etiology and autoimmune nature. The group of target polymorphisms was taken from our previous studies in identification of specific markers associated with virus hepatitis B and C. Similar to the method used in the previous study, the method of polymorphism detection was based on detection of SNP alleles using allele-specific locked nucleic acid (LNA) probes detectable in two or four channels of fluorescence detection [4]. The results were verified through Sanger sequencing. The comparative analysis of the allele distribution for the studied SNPs was performed using the Ensembl Genome Database Project [5] and the integrated map of the 1000 Genomes Project. The comparison was made using the distribution of frequencies of allelic variants for CEU subpopulations (Utah Residents with Northern and Western European Ancestry), FIN subpopulations (Finnish in Finland) and the EUR population (European). For the statistical analysis we used the standard fourfold table and the Pearson chi square test.
The study was conducted with the informed consent of the study participants. The study was approved by the Local Ethics Committee of the Central Research Institute of Epidemiology of Rospotrebnadzor (Protocol No. 114 dated April 22, 2021).
Results
The comparative analysis of the distribution of SNP allele frequencies, which was calculated for the group of relatively healthy population, and the data available in the Ensembl online resource demonstrated the following results: SNPs rs4374383 (MERTK), rs1143627 (IL1B), rs8099917 and rs12979860 (IL-28), rs28929474 (SERPINA1) did not have significantly significant differences with the compared groups. The highest similarity of allele frequency distributions for the studied polymorphisms was demonstrated by the EUR general population. SNPs rs1143634 (IL1B), rs1800795 (IL6), rs1800896 (IL10), and rs4251961(IL-1RN) had higher similarity with the FIN subpopulation, while the frequencies of polymorphisms rs419598 (IL1RN), rs9380516 (TULP1) and rs2295119 (HLA-DPA2) demonstrated high similarity with the frequencies of alleles typical of the CEU subgroup. At the same time, there were critical points showing significant differences between groups (p < 0.05). For example, frequencies of alleles of SNPs rs4986790 in the TLR4 gene differed significantly from the frequencies in the EUR population (p = 0.032) and in the CEU subpopulation (p = 0.047). Frequencies of polymorphisms rs1800795 (IL6) and rs1800896 (IL10) also significantly differed in the CEU subgroup (p = 0.030 and 0.012, respectively). It should also be noted that polymorphism rs2295119 (HLADPA2) had significant differences in frequencies in the EUR general population and almost the same level of differences in the FIN subpopulation. The summarized data are given in Table.
Table. Comparative characteristics of the distribution of polymorphism allele frequencies in the population of the Moscow region with the population of Europe
Таблица. Сравнительная характеристика распределения частот аллелей полиморфизмов населения Московского региона с населением Европы
SNP (rs) ID, gene Индивидуальный номер ОНП (rs), ген | Allele Аллель | Conditionally healthy population Условно здоровое население | 1000 GENOMES (EUR) | 1000 GENOMES (CEU) | 1000 GENOMES (FIN) | |||||||
n, abs. n, абс. | allele frequency частота аллеля | n, abs. n, абс. | allele frequency частота аллеля | p | n, abs. n, абс. | allele frequency частота аллеля | p | n, abs.. n, абс | allele frequency частота аллеля | p | ||
rs4986790 TLR4 | A | 871 | 0,92 | 949 | 0,943 | 0,032 | 190 | 0,96 | 0,047 | 175 | 0,884 | 0,114 |
G | 77 | 0,08 | 57 | 0,057 | 8 | 0,04 | 23 | 0,116 | ||||
rs4374383 MERTK | G | 1020 | 0,6 | 627 | 0,623 | 0,261 | 130 | 0,657 | 0,133 | 131 | 0,662 | 0,101 |
A | 676 | 0,4 | 379 | 0,377 | 68 | 0,343 | 67 | 0,338 | ||||
rs1143634 IL1B | G | 1550 | 0,76 | 757 | 0,752 | 0,594 | 152 | 0,768 | 0,841 | 151 | 0,763 | 0,967 |
A | 486 | 0,24 | 249 | 0,248 | 46 | 0,232 | 47 | 0,237 | ||||
rs1143627 IL-1B | A | 1321 | 0,65 | 652 | 0,648 | 0,942 | 128 | 0,646 | 0,622 | 123 | 0,621 | 0,428 |
G | 713 | 0,35 | 354 | 0,352 | 70 | 0,354 | 75 | 0,379 | ||||
rs4251961 IL1RN | T | 1356 | 0,67 | 639 | 0,635 | 0,086 | 123 | 0,621 | 0,197 | 140 | 0,707 | 0,249 |
C | 678 | 0,33 | 367 | 0,365 | 75 | 0,379 | 58 | 0,293 | ||||
rs419598 IL1RN | T | 1380 | 0,7 | 712 | 0,708 | 0,743 | 139 | 0,702 | 0,998 | 138 | 0,697 | 0,885 |
C | 586 | 0,3 | 294 | 0,292 | 59 | 0,298 | 60 | 0,303 | ||||
rs12979860 IFNL4 | C | 309 | 0,67 | 695 | 0,691 | 0,465 | 144 | 0,727 | 0,309 | 145 | 0,732 | 0,246 |
T | 151 | 0,33 | 311 | 0,309 | 54 | 0,273 | 53 | 0,268 | ||||
rs8099917 IFNL4 | T | 264 | 0,82 | 837 | 0,832 | 0,772 | 168 | 0,848 | 0,486 | 172 | 0,869 | 0,186 |
G | 56 | 0,18 | 169 | 0,168 | 30 | 0,152 | 26 | 0,131 | ||||
rs1800795 IL6 | G | 963 | 0,57 | 588 | 0,584 | 0,343 | 96 | 0,485 | 0,030 | 108 | 0,545 | 0,585 |
C | 739 | 0,43 | 418 | 0,416 | 102 | 0,515 | 90 | 0,455 | ||||
rs1800896 IL10 | T | 1166 | 0,57 | 550 | 0,547 | 0,175 | 95 | 0,48 | 0,012 | 119 | 0,601 | 0,442 |
C | 870 | 0,43 | 456 | 0,453 | 103 | 0,52 | 79 | 0,399 | ||||
rs28929474 SERPINA1 | C | 453 | 0,98 | 989 | 0,983 | 0,814 | 192 | 0,97 | 0,203 | 193 | 0,975 | 0,378 |
T | 7 | 0,02 | 17 | 0,017 | 6 | 0,03 | 5 | 0,025 | ||||
rs9380516 TULP1 | C | 1599 | 0,79 | 814 | 0,809 | 0,154 | 163 | 0,823 | 0,231 | 164 | 0,828 | 0,173 |
T | 433 | 0,21 | 192 | 0,191 | 35 | 0,177 | 34 | 0,172 | ||||
rs2295119 HLA-DPA2 | G | 684 | 0,85 | 888 | 0,883 | 0,034 | 175 | 0,884 | 0,207 | 178 | 0,899 | 0,069 |
T | 122 | 0,15 | 118 | 0,117 | 23 | 0,116 | 20 | 0,101 | ||||
Note. Statistically significant differences are shown in bold.
Примечание. Жирным шрифтом выделены статистически значимые отличия в сравниваемых группах.
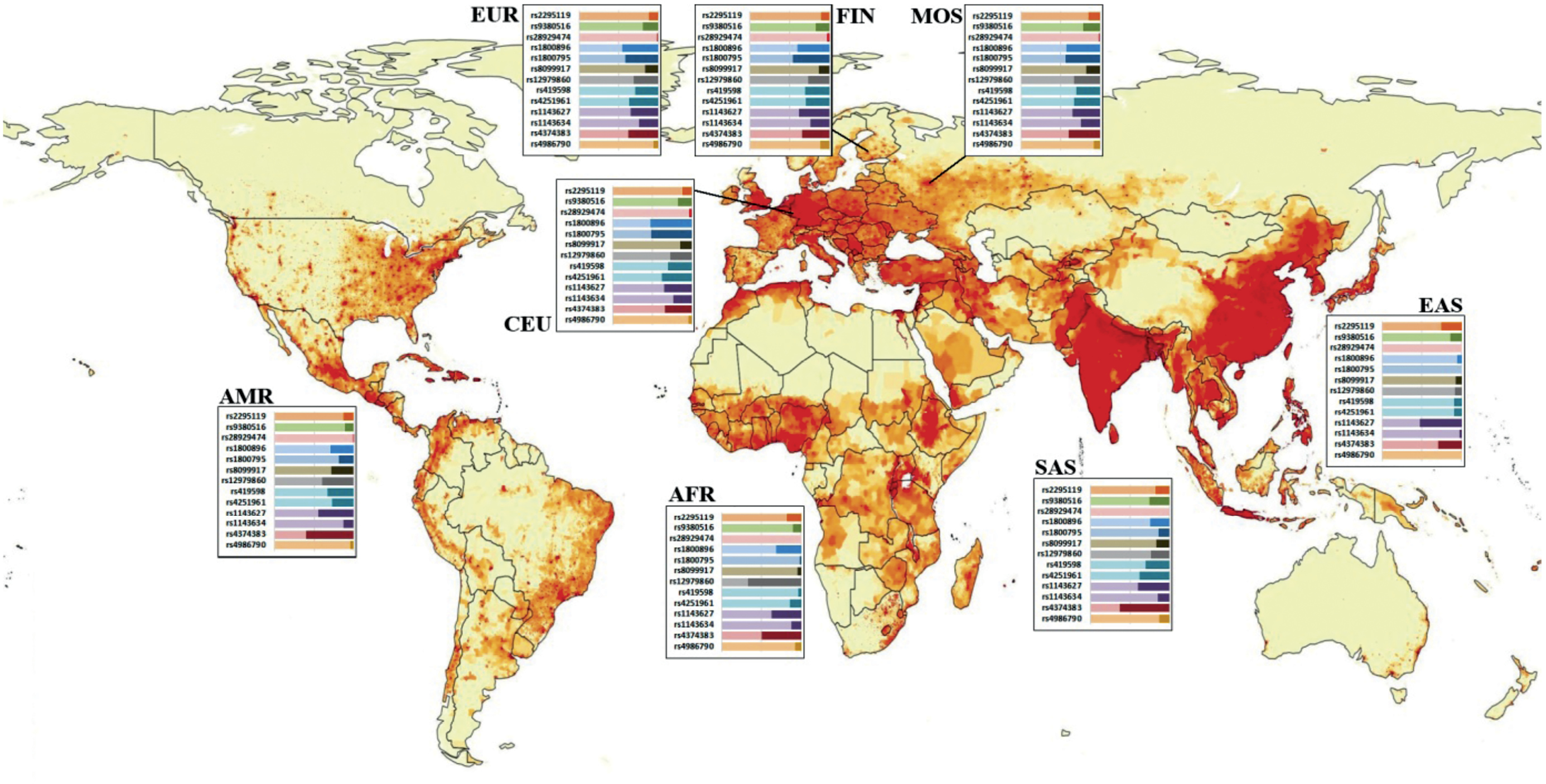
The figure shows the distribution of allele frequencies among representatives of 5 major world populations, two FIN and CEU subpopulations that are most closely related to the population of the European part of Russia, and the MOS group having its own characteristics identified for the relatively healthy population of the Moscow region.
As can be seen in the figure, each polymorphism, to a certain extent, has a different distribution of allelic variants among major world populations. Statistically significant differences between populations and subpopulations deserve special attention. The alternative allele of polymorphism rs2295119 (HLADPA2) (G/T) is detected within the 12 to 26% range among the world populations, thus providing excellent possibilities for exploring the role of this genetic variant in various pathological conditions. For the MOS cohort, the frequency of occurrence of the alternative allele among the participants of this study significantly differed from the frequencies in the EUR population and was approaching the values that would make it significantly different from the frequencies in the FIN subpopulation.
SNPs rs9380516 (TULP1) demonstrated the highest heterogeneity in the SAS population (25%); however, this polymorphism is characterized by average high allele frequencies. The AMR and AFR populations demonstrated the lowest frequencies of the alternative allele, accounting for 11%.
SNP rs1800795 (IL6) had a wide range of alternative allele frequencies: from 0% in the EAS population to 52% in the CEU population. Note that based on the data from the frequency distribution databases, the EAS population, which includes representatives of such countries as China, Vietnam, and Japan, is totally homogenous by the prevalence of the reference allele (G). However, quite a few studies have found the association of this polymorphism with pathologies of different etiology among Asians [6–8]. Several meta-analyses have been conducted, focusing on research findings, including the results obtained for the regions, the population of which does not have allelic heterogeneity for polymorphism rs1800795 according to the Ensembl database [9, 10].
The coefficient of determination, R2, shows the level of linkage of heritable characteristics; its value ranges from 0 to 1. The linkage of genetic polymorphisms directly depends on their chromosomal location and on other factors. Identification of linkage groups is of critical importance in evaluation of the association between SNPs and the pattern of pathological processes. In the meantime, the existing international databases are notorious for controversies regarding the same SNPs. In our study, we used several pairs of polymorphisms belonging to the same gene. For example, the pair of well-studied polymorphisms (rs8099917 and rs12979860) belonging to the gene encoding interferon-λ4 (IFN-λ4), according to the LDlink resource [11] based on the international dbSNP database, has a wide spread of R2 correlation coefficient values – from the complete absence of correlation in the AFR population (R2 = 0.02) to the insignificant correlation of alleles in the EUR population (R2 = 0.04) and almost 100% correlation in the EAS population (R2 = 0.98).
SNPs rs419598 and rs4251961 of the IL-1 receptor antagonist gene (IL1RN) are inherited independently of each other in all world populations, having the highest values – R2 = 0.24 in the SAS population and R2 = 0.23 in the EUR population. The frequency of the alternative allele of SNP rs4251961 varies significantly across populations. The lowest frequency rate is detected in the EAS population, being 9%, while the SAS and EUR populations demonstrate the highest allelic heterogeneity of these loci with the frequency rates of the alternative allele reaching 38% and 36%, respectively. For SNP rs419598, the alternative allele frequency range is also wide, from 3% in the AFR population to 33% – in AMR. In the European population (EUR), the alternative allele is found in 29% of the population.
Polymorphisms of the IL1B gene (rs1143627 and rs1143634) are inherited independently of each other. The generalized R2 coefficient for all the populations is 0.03. The alternative allele of SNP rs1143634 is detected at the frequency rate ranging from 2% in the EAS population to 25% in the EUR population. The distribution of allele frequencies for polymorphism rs1143627 also varies significantly among world populations; the lowest frequency rate is 35% in the EUR population, reaching 52% in the EAS population.
The pair of SNPs rs8099917 and rs12979860 belongs to the gene encoding IFN-λ3. Based on the LD pair tool, the R2 coefficient for this pair of SNPs demonstrates a wide spread in values: the complete absence of correlation between two genetic traits in the AFR population (R2 = 0.02), insignificant statistical allelic correlation in the EUR population (R2 = 0.04) and almost 100% correlation in the EAS population (R2 = 0.98). Currently, this phenomenon cannot be explained and requires further thorough research.
Polymorphism rs28929474 of the SERPINA1 gene presents an example of nearly 100% genetic homogeneity in the world population. According to the Ensebl database, the variability of this locus was found only in EUR and AMR populations; the alternative allele is found only in 2% and 0.4% of the population, respectively. The breakdown of the EUR population into subpopulations reveals an interesting pattern: The alternative allele frequency rate in the CEU subpopulation is 3%, in FIN – 2.5%, GBR – 1.1%, IBS – 1.9%, and TCI – 0%, while the MOS group has 2% typical of the EUR population. This polymorphic locus has only two possible structural variants: heterozygous with cytosine and thymine (CT) and homozygous with cytosine (CС). The SERPINA1 gene encodes α-1-antitrypsin – a serine protease inhibitor (serpin). The polymorphic locus rs28929474 is generally studied in combination with the second polymorphism of the SERPINA1 gene, namely rs17580. It has been found that the homozygous combination of the above pair of polymorphisms, the PiZZ variant, is the most common cause of severe α-1-antitrypsin deficiency and associated pathological conditions such as pulmonary pathologies, liver cirrhosis, hepatocellular carcinoma, vasculitis, and panniculitis [12].
Discussion
The expansion of molecular biology methods opened new opportunities in exploration of the gene structure and, consequently, in identification of structural variants of coding and regulatory regions of human genes. The Human Genome Project, one of the greatest scientific feats in history, laid the groundwork for numerous studies focused on the role of genes in the development of diseases and addressing other scientific problems in the field of medicine. The 1000 Genomes Project, one of the most successful large-scale projects in human genetics, was completed in the 2000s [13]. It created a publicly available reference database that is actively used for analysis of the sequence data obtained in different studies. The development of molecular research methods and the increased availability of sequencing paved the way for multiple resources in genetics of humans, animals, insects, and plants. Through joint efforts, such projects as ALPHA, HapMap and others contribute to the knowledge of the genetic diversity of the human population; however, the information collected by these projects cannot provide the comprehensive coverage of the genetic diversity of the world population. Currently, the international databases have data on 5 major populations: African (AFR), American (AMR), East Asian (EAS), European (EUR), and South Asian (SAS). Each of them is additionally divided into subpopulations. The European population is represented by 5 subpopulations: CEU (Utah Residents with Northern and Western European Ancestry), FIN (Finnish in Finland), GBR (British in England and Scotland), IBS (Iberian populations in Spain), and TSI (Toscani in Italy). In their studies in regions that are left out from the human genetics databases, for example, in Russia, researchers use the most closely related subpopulations as reference. Our study has demonstrated that some SNPs typed in the cohort of the population of the European part of Russia have the highest homology with the subpopulation of indigenous people of Finland (FIN), while some SNPs have a higher homology with the EUR population by their detection frequency rates. This situation limits the use of the existing international databases in studies exploring the association of SNPs with various pathological conditions. At the moment, the results of studies conducted by Russian research groups [14] cannot be combined into a single information resource aggregating data on the so-called basic distribution of frequencies of SNPs alleles and genotypes among the population of Russia. In the meantime, considering the current level of development of science, creation of such database is a top-priority task.
The reliability of medical and biological studies is a critical parameter in searching for genetic determinants affecting various aspects of the pathological process, including its initial stage, development variants, complications and outcomes of infectious and somatic diseases. At the same time, scientific publications addressing the above problem can have controversial information due to the lack of relevant reference data. Comparison of the newly identified polymorphism frequencies with international databases is also affected by small sizes of sample subsets for some subpopulations. For example, for SNP rs1800795 (IL6) in CEU and FIN subpopulations, the Ensembl online resource has data on distribution of allele frequencies for 198 people and data on genotypes for the subset composed of 99 participants – the sizes being critically insufficient for the comparative analysis [15]. The analysis of the scientific literature in the discussed field showed that most of the studies were performed using small-sized subsets both as groups of patients having the studied characteristics and as comparison groups, which usually included not more than 150 people [16–18]. The above size (150 people) cannot ensure sufficient accuracy in most of the studies. Fox et al. [19] offered a reference chart specifying the error margin and the respective sample size. Based on their criteria, if the sample error is 5%, the recommended size of the group must be at least 350 people. Compliance with the representativeness requirements for study and control groups is mandatory to support the reliability of studies in the target field. Urgent efforts are required to resolve the problem of the absence of its own information system in Russia, which would contain the data incorporating racial and ethnic diversity of population.
Figure. The ratio of allele frequencies of a number of polymorphisms among the main populations of the world.
The data are presented in the Ensembl international database. MOS – own data obtained on the territory of the Moscow region. The basis for the image was taken from the resource https://worldinmaps.com/ world/population-and-settlement/population-density/
Рисунок. Соотношение частот аллелей ряда полиморфизмов среди основных популяций мира.
Данные представлены в международных базах данных ресурса Ensembl. MOS – собственные данные, полученные на территории Московского региона. Основа для изображения взята с ресурса https://worldinmaps.com/world/population-and-settlement/population-density/
This study offers compelling evidence of significant genetic heterogeneity of the population, even within limited areas; for clinical and preventive medical studies, such high heterogeneity necessitates the need for a deep insight into what we define as “the genetic profile of the population”. Many present-day medical and biological studies focus on promotion of a personalized approach to the patient in healthcare and, consequently, on identification of genetic markers indicating possible risks of development of a pathological condition. At the same time, the results of such studies can be valuable as monitoring parameters in epidemiological surveillance of infectious diseases. Mathematical models, which are built to predict incidence rates, mortality, complication rates, and other monitoring parameters characterizing the epidemic process, must incorporate the maximum possible number of factors, including genetic determinants of the population. However, such calculations require continuous identification of new genetic determinants and monitoring of allele frequencies of marker genes. At present, Russian scientists do not have sufficient data on allele prevalence for human marker genes to develop efficient mathematical models, thus being in need for activization and expansion of respective scientific studies.
Reliable results of genetic and statistical studies are extremely difficult to receive, considering the differences in distribution of allele frequencies for polymorphisms of human genes. The results of the studies conducted in Russia in compliance with the requirements for the selection of target and comparison groups can be of low significance, as there is no confirmation that the comparison group represents the average values of allele frequencies typical of the relatively healthy population. Note that the cohort selected as a comparison group can match the allele frequencies available in international databases [20]. The increased size of the sample subset demonstrates the discrepancy in population-wide distribution of allele frequencies, thus suggesting that the homology between the data from the comparison group and the reference data is of random nature. Infectious diseases such as virus hepatitis, HIV-associated immunodeficiency and others are intently studied by the scientific community. Different disease outcomes are associated with the genetic determinants responsible for human immune responses to the pathogen. The search for specific genetic markers serving as predictors of the outcome and the host response to the administered treatment requires a group with the studied characteristics (the study group) and without them (the comparison group). The discrepancy between the data of the comparison group and the general population data can produce false-positive associations with conditions caused by viral agents.
Conclusion
The existing data subsets, though coming from worldwide locations, are fragmentary, as they present characteristics of local populations. The small number of participants in studies is another important factor contributing to the unreliability of the data on gene variants and, therefore, to their unsuitability for mathematic modeling of the SNP impact on development of epidemic processes within a specific population. Statistically significant data can be produced when supported by a national information system containing information about the genetic profile of the population of regions in Russia, and this system must be available to research institutions both for its expansion through adding new data and for its use in respective mathematical models.
The comparison of SNP frequency distribution in relatively healthy population with the reference data of the international Ensembl resource highlights the need for the Russia-based information system containing data on the occurrence of SNP alleles and genotypes among relatively healthy population and in subgroups with different pathological conditions. The analysis revealed significant differences in frequencies of some polymorphisms in the MOS group compared to reference frequencies from international databases as well as the inaccuracy of the reference data on the distribution of allele frequencies in major populations using polymorphism rs1800795 of the IL6 gene and publications of Chinese researchers providing multiple examples of discrepancy in reference data. The unavailability of reliable information about most of the regions in Russia demonstrates the significance of a national database giving comprehensive information coverage of the genetic diversity of the population of the country. Reference data incorporating racial and ethnic diversity of the population of Russia will enhance the reliability of study results in identification of specific markers affecting the development of pathological conditions in humans.
About the authors
Natalia V. Vlasenko
Central Research Institute for Epidemiology, Federal Service for Surveillance on Consumer Rights Protection and Human Wellbeing
Author for correspondence.
Email: nvzuz@mail.ru
ORCID iD: 0000-0002-2388-1483
Researcher, Laboratory of viral hepatitis
Russian Federation, 111123, MoscowMikhail D. Chanyshev
Central Research Institute of Epidemiology, Federal Service for Surveillance on Consumer Rights Protection and Human Wellbeing
Email: chanish@mail.ru
ORCID iD: 0000-0002-6943-2915
Researcher, Laboratory of genomic research
Russian Federation, 111123, MoscowDmitriy V. Dubodelov
Central Research Institute for Epidemiology, Federal Service for Surveillance on Consumer Rights Protection and Human Wellbeing
Email: gradient27@mail.ru
ORCID iD: 0000-0003-3093-5731
Cand. Sci. (Med.), senior researcher, Laboratory of viral hepatitis
Russian Federation, 111123, MoscowArtem A. Serkov
Central Research Institute for Epidemiology, Federal Service for Surveillance on Consumer Rights Protection and Human Wellbeing
Email: toms8191@mail.ru
ORCID iD: 0009-0006-4086-8324
Research laboratory assistant, Laboratory of viral hepatitis
Russian Federation, 111123, MoscowGalina G. Solopova
Dmitry Rogachev National Medical Research Center of Pediatric Hematology
Email: galina.solopova@fnkc.ru
ORCID iD: 0000-0002-1680-7269
PhD, Deputy Chief Physician for Infection Control
Russian Federation, 117997, MoscowAnastasija V. Sacuk
Dmitry Rogachev National Medical Research Center of Pediatric Hematology
Email: vnpoemp@yandex.ru
ORCID iD: 0000-0003-3293-2008
PhD, epidemiologist
Russian Federation, 117997, MoscowArtem V. Snicar
Demikhov Moscow City Clinical Hospital
Email: snitsarav@yandex.ru
ORCID iD: 0000-0001-6053-4651
Deputy Chief Medical Officer
Russian Federation, 109263, MoscowTatiana А. Semenenko
N.F. Gamaleya National Research Center of Epidemiology and Microbiology
Email: semenenko@gamaleya.org
ORCID iD: 0000-0002-6686-9011
D. Sci. (Med.), Professor, Head of the Epidemiology Department
Russian Federation, 123098, MoscowStanislav N. Kuzin
Central Research Institute for Epidemiology, Federal Service for Surveillance on Consumer Rights Protection and Human Wellbeing
Email: drkuzin@list.ru
ORCID iD: 0000-0002-0616-9777
D. Sci. (Med.), Professor, Head, Laboratory of viral hepatitis
Russian Federation, 111123, MoscowVasily G. Akimkin
Central Research Institute for Epidemiology, Federal Service for Surveillance on Consumer Rights Protection and Human Wellbeing
Email: crie@pcr.ru
ORCID iD: 0000-0003-4228-9044
D. Sci. (Med.), Professor, Full Member of the Russian Academy of Sciences, Director
Russian Federation, 111123, MoscowReferences
- Kuper L., ed. Race, Science and Society. Paris: The Unesco Press; 1975.
- Rychkov Yu.G., ed. Gene Pool and Genogeography of the Population. Volume I. The Gene Pool of the Population of Russia and Neighboring Countries [Genofond i genogeografiya narodonaseleniya. Tom 1. Genofond naseleniya Rossii i sopredel’nykh stran]. St. Petersburg: Nauka; 2000. (in Russian)
- Likhvantsev V.V., Yadgarov M.Ya., Berikashvili L.B., Kadantseva K.K., Kuzovlev A.N. Sample size estimation. Anesteziologiya i reanimatologiya. 2020; (6): 7786. https://doi.org/10.17116/anaesthesiology202006177 (in Russian)
- Dribnokhodova O.P., Korchagin V.I., Mironov K.O., Dunaeva E.A., Titkov A.V., Aksel’rod E.V., et al. A comparative analysis of allele frequencies of rs1801133 and rs1801131 of MTHFR in patients with stroke and healthy people from the Moscow region. Zhurnal nevrologii i psikhiatrii im. S.S. Korsakova. 2019; 119(3-2): 18–23. https://doi.org/10.17116/jnevro201911903218 https://elibrary.ru/mfftvn (in Russian)
- Ensembl genome browser 110. Available at: https://www.ensembl.org/index.html
- Yin Y.W., Sun Q.Q., Zhang B.B., Hu A.M., Wang Q., Liu H.L., et al. The lack of association between interleukin-6 gene -174 G/C polymorphism and the risk of type 1 diabetes mellitus: a meta-analysis of 18,152 subjects. Gene. 2013; 515(2): 461–5. https://doi.org/10.1016/j.gene.2012.11.062
- Xia J., Sun R.L. Association between interleukin-6 rs1800795 polymorphism and the decreased risk of type 2 diabetes mellitus: an updated meta-analysis. Int. J. Clin. Exp. Med. 2019; 12(1): 86–97.
- Zhang X., Ma L., Peng F., Wu Y., Chen Y., Yu L., et al. The endothelial dysfunction in patients with type 2 diabetes mellitus is associated with IL-6 gene promoter polymorphism in Chinese population. Endocrine. 2011;40(1):124–9. https://doi.org/10.1007/s12020-011-9442-9
- Yang Y., Xiao J., Tang L., Wang B., Sun X., Xu Z., et al. Effects of IL-6 polymorphisms on individual susceptibility to allergic diseases: a systematic review and meta-analysis. Front. Genet. 2022; 13: 822091. https://doi.org/10.3389/fgene.2022.822091
- Cheng Z., Zhang C., Mi Y. IL-6 gene rs1800795 polymorphism and diabetes mellitus: a comprehensive analysis involving 42,150 participants from a meta-analysis. Diabetol. Metab. Syndr. 2022; 14(1): 95. https://doi.org/10.1186/s13098-022-00851-8
- National Institutes of Health. LDlink. Available at: https://ldlink.nci.nih.gov/
- Diagnosis and treatment of pulmonarydisease in α1-antitrypsin deficiency: a statement of European Respiratory Society. Pul’monologiya. 2018; 28(3): 273–95. https://doi.org/10.18093/0869-0189-2018-28-3-273-295 (in Russian)
- Risch N.J. Searching for genetic determinants in the new millennium. Nature. 2000; 405(6788): 847–56. https://doi.org/10.1038/35015718
- Voevoda M.I. Polymorphysm and connection with risk factors of some genes of predisposition to cardiovascular diseases in ethnic groups in Siberia (molecular-epidemiological and evolution-genetic aspects). Ateroskleroz. 2009; 5(1): 3–27. https://elibrary.ru/nxqejt (in Russiam)
- Ensembl genome browser 110. rs1800795 SNP – Explore this variant – Homo_sapiens. Available at: https://www.ensembl.org/Homo_sapiens/Variation/Explore?r=7:22726526-22727526;v=rs1800795;vdb=variation;vf=729516845
- Zotova I.I., Kapustin S.I., Gritsaev S.V., Mineeva N.V., Krobinets I.I., Sidorova Zh.Yu., et al. GPIIB allelic polymorphism as a factor associated with the probability of immune thrombocytopenia and the severity of hemorrhagic syndrome. Onkogematologiya. 2018; 13(2): 93–9. https://doi.org/10.17650/1818-8346-2018-13-2-93-99 https://elibrary.ru/uuawru (in Russian)
- El’kina A.Yu., Akimova N.S., Shvarts Yu.G., Martynovich T.V., Fedotov E.A. Vascular control parameters and gene polymorphism associated with cardiovascular risk in young and relatively healthy individuals. Kardiovaskulyarnaya terapiya i profilaktika. 2019; 18(2): 45–50. https://doi.org/10.15829/1728-8800-2019-2-45-50 https://elibrary.ru/zclfjj (in Russian)
- Belokrinitskaya T.E., Frolova N.I., Strambovskaya N.N. Gene polymorphism associated with risk of development of homocysteine exchange disorders among young healthy women in Transbaikal Kray: ethnic and reproductive aspects. Byulleten’ Vostochno-Sibirskogo nauchnogo tsentra Sibirskogo otdeleniya Rossiyskoy akademii meditsinskikh nauk. 2013; (5): 13–6. https://elibrary.ru/rtuewh (in Russian)
- Fox N., Hunn A., Mathers N. Sampling and sample size calculation. In: The NIHR RDS for the East Midlands. Yorkshire and the Humber; 2009
- Vlasenko N.V., Churilova N.S., Loskutova T.A., Mironov K.O., Es’man A.S., Dunaeva E.A., et al. Evaluation of the epidemiological significance of molecular genetic factors in relation to the intensity of post-vaccination immunity against hepatitis B. Zhurnal mikrobiologii, epidemiologii i immunobiologii. 2022; 99(2): 150–59. https://doi.org/10.36233/0372-9311-246 https://elibrary.ru/mtosqh (in Russian)
Supplementary files