Genetic characteristics of the isolate of human adenovirus type 55 (Adenoviridae: Mastadenovirus) isolated in Moscow in 2022
- Authors: Shein D.A.1, Ryzhova N.N.1, Kunda M.S.1, Ermolova E.I.1, Ozharovskaia T.A.1, Popova O.1, Nikitenko N.A.1, Krasnoslobodtsev K.G.1, Burtseva E.I.1, Zubkova O.V.1, Voronina O.L.1, Gintsburg A.L.1
-
Affiliations:
- National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya of the Ministry of Health of the Russian Federation
- Issue: Vol 70, No 5 (2025)
- Pages: 431-443
- Section: ORIGINAL RESEARCHES
- URL: https://virusjour.crie.ru/jour/article/view/16721
- DOI: https://doi.org/10.36233/0507-4088-297
- EDN: https://elibrary.ru/aatdbs
- ID: 16721
Cite item
Abstract
Introduction. Adenovirus infection occurs globally in the form of sporadic cases and isolated outbreaks. Human adenovirus type 55 (HAdV-55), endemic in China and South Korea, causes acute respiratory viral infections (ARVI) of varying severity, both among the civilian population and in military units in different countries of the world. Genomic research facilitates reliable identification of HAdV-55.
The aim of this study was to identify HAdV isolated in Moscow in 2022, as well as to conduct whole-genome sequencing and comparative genomic research.
Materials and methods. HAdV-55 was isolated from a sample of a patient hospitalized with pneumonia and analyzed using restriction fragment length polymorphism analysis and whole-genome sequencing. Bioinformatics comparative analysis was performed on a sample of sequences of 83 isolates.
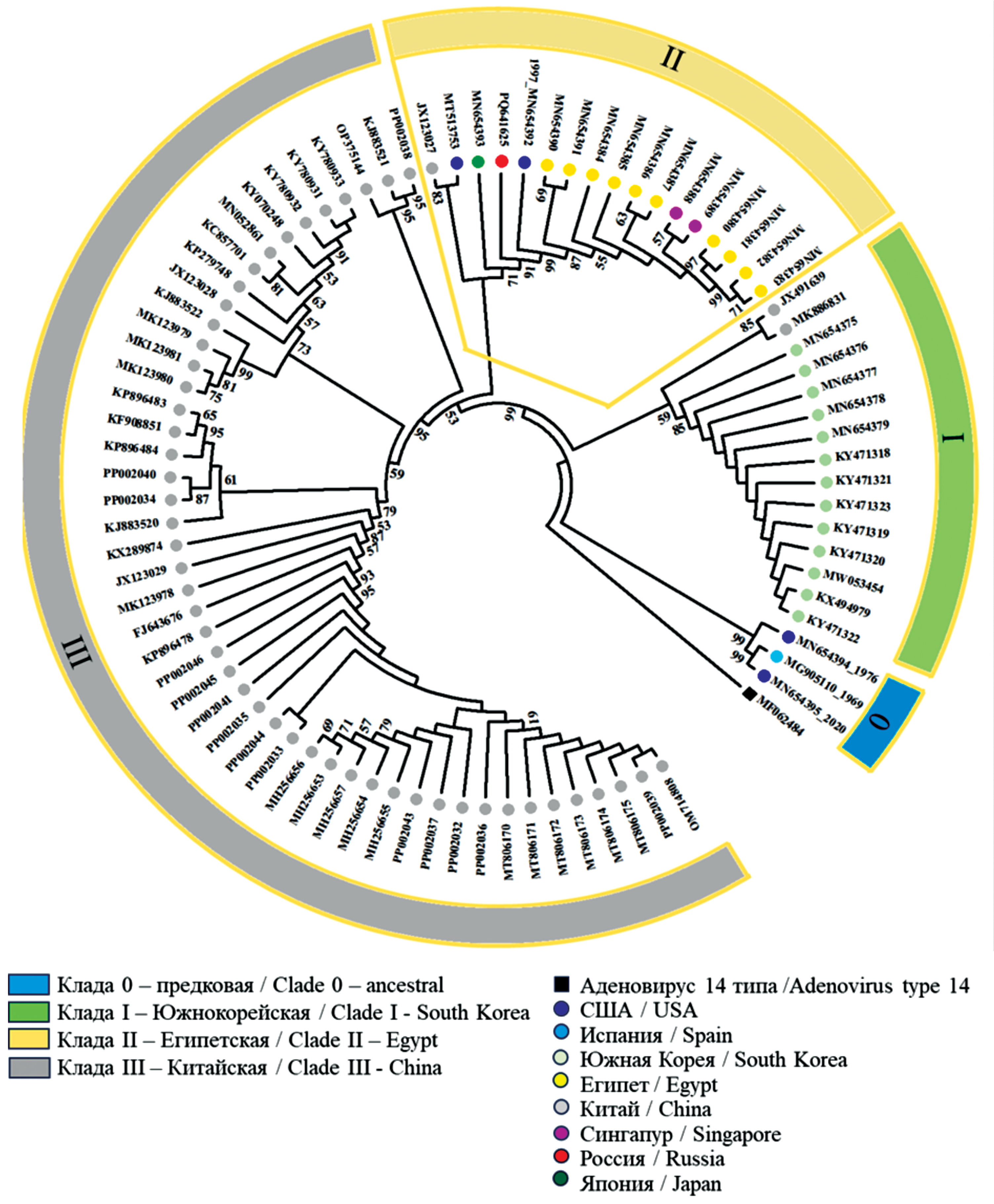
Results. The whole-genome sequencing of first isolated in Russia HAdV-55 was conducted. The sequence of isolate SCV3008:Ad55 was deposited in GenBank (Accession Number PQ641625). Unique mutations in the SCV3008:Ad55 genome were identified, one of which resulted in a conservative T29A substitution in the penton that did not affect its functions. Phylogenetic analysis showed clustering of SCV3008:Ad55 with isolates of clade II, which included representatives of 7 countries on different continents, indicating a wide distribution of HAdV-55. Isolates from endemic regions of China and South Korea formed separate clades. The study of microsatellite length polymorphism in untranslated regions of the genome became an additional tool for distinguishing closely related genomes.
Conclusion. The obtained genomic information laid the foundation for further monitoring for HAdV-55 in Russia and demonstrated the informativeness and significance of whole-genome studies for monitoring adenoviruses. The development and implementation of genotyping methods aimed at detecting HAdV-55 and other clinically relevant genotypes will significantly improve the effectiveness of the diagnosis of adenovirus infections with the threat of developing bronchopneumonia.
Full Text
Introduction
Adenoviruses (family Adenoviridae) are non-enveloped, double-stranded DNA viruses, classified into 6 genera: Aviadenovirus, Barthadenovirus, Ichtadenovirus, Mastadenovirus, Siadenovirus and Testadenovirus. Mammalian adenoviruses belong to the genus Mastadenovirus, which includes more than 50 species. Human adenoviruses (HAdV) belong to 7 species: Mastadenovirus adami, M. blackbeardi, M. caesari, M. dominans, M. exoticum, M. faecale and M. russelli1. Different types of HAdV have varying tissue tropism, which often correlates with specific clinical symptoms of the infection [1]. HAdV primarily cause acute respiratory viral infections (ARVI), but can also affect the eyes, intestines, urinary tract and nervous system. The severity of the disease depends on the type of virus and the host’s immune status [1–5]. The most severe respiratory infections are caused by HAdV 8 genotypes out of 10 belonging to the M. blackbeardi species: 3, 7, 11, 14, 16, 21, 50, 55 [6].
The history of isolating HAdV-55 as a separate genotype demonstrates the role and development of the methodological base of virology, contributing to a more accurate classification of viruses. For the first time, the atypical HAdV-11 virus as a pathogen of ARVI was identified using immunochemical methods in 1974 [7]. In 1991, the comparison of DNA fragment length polymorphism of isolates allowed the identification of the HAdV-11a genotype and demonstrated that viruses of this genotype are associated with upper respiratory tract infections and bronchopneumonia [8]. In 2009, the first complete genome of the HAdV-11a isolate HAdV11-QS (registration number FJ643676) was published, obtained by assembling overlapping amplicons after Sanger sequencing. Comparison of sequencing data contributed to the demonstration of the origin of HAdV-11a through recombination: the HAdV-11a genome is based on the HAdV-14 genome and part of the hexon gene of HAdV-11 [9]. In 2011, the Human Adenovirus Working Group recommended using full-genome sequences for typing and characterizing HAdV and classifying recombinants into new genotypes based on differences in nucleotide sequences and biological properties [10]. Based on these recommendations, in 2013, the recombinant HAdV-11a was named HAdV-55 with the prototype isolate HAdV11-QS [11]. Retrospective studies of collection isolates have shown that HAdV-55 is endemic to China and South Korea and dominated among viruses isolated during ARVI in Beijing from 1965 to 1985 [8]. Outbreaks of HAdV-55 ARVI in organized groups have been recorded since 1969 among military personnel in Spain [7], in a vocational training center in the USA [12], among children in Argentina, Chile and Uruguay [13], in psychiatric institutions in Israel [14], and in family groups in China [15]. HAdV-55, compared to respiratory adenoviruses of other genotypes, causes more severe diseases and poses a significant threat to public health [6]. In Russia, according to data from the Center for Ecology and Epidemiology of Influenza at the D.I. Ivanovsky Institute of Virology, N.F. Gamaleya NRCEM of the Ministry of Health of Russia, in collaboration with 10 reference bases, during the epidemic season of 2021–2022, the frequency of positive adenovirus samples in the studied areas was 7.7% (1793 samples) out of those tested for ARVI [16]. HAdV, isolated from one of the 1793 samples, became the subject of detailed study.
The aim of the study is the identification of HAdV isolated in Moscow in 2022, as well as conducting whole-genome sequencing and comparative genomic study.
Materials and methods
Materials. Bronchoalveolar lavage (BAL) from a hospitalized 34-year-old man with a diagnosis of unspecified pneumonia. The study was conducted with the voluntary informed consent of patients. The study protocol was approved by the Ethics Committee of the Moscow State Medical University «Infectious Clinical Hospital No. 1 of the Moscow Department of Health» (Protocol No. 8 dated 12/28/2022).
Methods. The identification of viruses was carried out by extracting RNA/DNA from clinical material using the RIBO-PREP kit (Interlabservice, Russia) followed by the detection of RNA/DNA of respiratory infection pathogens in real-time reverse transcription polymerase chain reaction (RT-PCR) using AmpliSens ARVI-screen-FL commercial test systems (Interlabservice, Russia) according to the manufacturer’s instructions on the Bio-Rad CFX-96 detection amplifier (Bio-Rad, USA).
Isolation of HAdV. The virus was accumulated in HEK293 cells (human embryonic kidney): 100 µL of BAL was used to infect the cells (0.5 × 106 cells/3 cm², incubated under standard conditions (+37 °C, 5% CO2) until cytopathic effect (CPE) occurred. For the preparative amplification of the virus, 15 cm diameter culture dishes were used. Infected cells were collected after reaching 100% CPE, concentrated by low-speed centrifugation (2000 rpm/10 min), re-suspended in buffer (0.01 M Tris-HCl pH 8.0, 0.01 M NaCl, 5 mM EDTA), subjected to three freeze-thaw cycles, and centrifuged at 5000 rpm/10 min, with the pellet being discarded. The adenovirus was purified from the supernatant using ultracentrifugation in a cesium chloride density gradient (in stepwise (CsCl with refractive indices of 1.355, 1.365, and 1.375) and equilibrium gradient (CsCl with a refractive index of 1.365)).
Analysis of restriction fragment length polymorphism (RFLP). Genomic DNA from the purified virus was extracted using the Wizard Genomic DNA Purification Kit (Promega, USA). DNA (1 µg) was hydrolyzed with the restriction enzymes Cfr41I, XagI and XhoI (Thermo, USA) and analyzed by agarose gel electrophoresis, using a longer incubation period to detect low molecular weight fragments. In silico analysis of RFLP was conducted using the Geneious Prime software (Biomatters, New Zealand).
Sequencing and genome assembly. Library preparation was performed using the KAPA HyperPlus Kit (F. Hoffmann-La Roche Ltd., Switzerland) according to the manufacturer’s protocols, quality and size checks of the libraries were conducted using electrophoresis on High Sensitivity DNA Chips 2100 Bioanalyzer System (Agilent, USA), sequencing was carried out on the NextSeq 500/550 (Illumina, USA) instrument using Mid Output 300 cycles cartridges. For de novo assembly and reference sequence assembly, we used the CLC Genomic Workbench v. 21 software package (Qiagen, USA). To refine the sequences of homopolymers, Sanger sequencing was performed, using the BDT UltraSeq HP Kit (SenseCare Bio, China), with electrophoresis conducted in 50 cm capillaries in POP-7 gel on the 3500 Genetic Analyzer (Applied Biosystems, USA).
Comparative analysis. The comparative analysis included 83 complete genomes of HAdV-55 (Table 1) and the genome of HAdV-14 (MF062484). The alignment of genomic sequences of the isolate samples, the construction of the Neighbor-joining phylogenetic tree, and the calculation of ANI (average nucleotide identity) were performed using the Whole Genome Alignment module of the CLC Genomic Workbench v. 21 software package (Qiagen, USA). The MEGA11 program was used for tree visualization, as well as translation of reading frames and alignment of amino acid sequences [17].
Table 1. HAdV-55 strains used for genomic analysis in order to study the spread of the virus, regional persistence and genetic variability
Таблица 1. Штаммы HAdV-55, используемые для геномного анализа с целью изучения распространения вируса, региональной персистенции и генетической изменчивости
GenBank Accession Number Номер в базе NCBI | Country Место выделения | Year of isolation Год выделения | GenBank Accession Number Номер в базе NCBI | Country Место выделения | Year of isolation Год выделения |
MN654381.1 | Egypt / Египет | 2000 | PP002035.1 | China / Китай | 2018 |
MN654383.1 | Egypt / Египет | 2000 | PP002036.1 | China / Китай | 2018 |
MN654385.1 | Egypt / Египет | 2000 | PP002037.1 | China / Китай | 2018 |
MN654380.1 | Egypt / Египет | 2000 | PP002043.1 | China / Китай | 2018 |
MN654382.1 | Egypt / Египет | 2002 | PP002044.1 | China / Китай | 2018 |
MN654390.1 | Egypt / Египет | 2005 | PP002045.1 | China / Китай | 2018 |
MN654391.1 | Egypt / Египет | 2005 | PP002046.1 | China / Китай | 2018 |
MN654386.1 | Egypt / Египет | 2007 | MH256653.1 | China / Китай | 2018 |
MN654384.1 | Egypt / Египет | 2008 | MH256655.1 | China / Китай | 2018 |
MN654387.1 | Egypt / Египет | 2009 | MH256657.1 | China / Китай | 2018 |
MG905110.1 | Spain / Испания | 1969 | MH256654.1 | China / Китай | 2018 |
FJ643676.1 | China / Китай | 2006 | MH256656.1 | China / Китай | 2018 |
JX123027.1 | China / Китай | 2010 | PP002040.1 | China / Китай | 2018 |
JX491639.1 | China / Китай | 2011 | MT806174.1 | China / Китай | 2019 |
JX123028.1 | China / Китай | 2011 | MT806175.1 | China / Китай | 2019 |
MK123979.1 | China / Китай | 2011 | MT806170.1 | China / Китай | 2019 |
KJ883522.1 | China / Китай | 2011 | MT806172.1 | China / Китай | 2019 |
KP279748.1 | China / Китай | 2012 | MT806173.1 | China / Китай | 2019 |
KP896478.1 | China / Китай | 2012 | MT806171.1 | China / Китай | 2019 |
JX123029.1 | China / Китай | 2012 | OM714808.1 | China / Китай | 2020 |
KC857701.1 | China / Китай | 2012 | OP375144.1 | China / Китай | 2021 |
KP896483.1 | China / Китай | 2013 | MN654388.1 | Singapore / Сингапур | 2005 |
KJ883520.1 | China / Китай | 2013 | MN654389.1 | Singapore / Сингапур | 2005 |
KJ883521.1 | China / Китай | 2013 | MN654394.1 | США / USA | 1976 |
KP896484.1 | China / Китай | 2013 | MN654392.1 | США / USA | 1997 |
MK123980.1 | China / Китай | 2013 | MT513753.1 | США / USA | 2006 |
MK123981.1 | China / Китай | 2013 | MN654395.1 | США / USA | 2020 |
KF908851.1 | China / Китай | 2013 | MN654375.1 | South Korea / Южная Корея | 2009 |
MK886831.1 | China / Китай | 2015 | MN654376.1 | South Korea / Южная Корея | 2009 |
KX289874.1 | China / Китай | 2015 | MN654377.1 | South Korea / Южная Корея | 2009 |
KY070248.1 | China / Китай | 2016 | MN654378.1 | South Korea / Южная Корея | 2009 |
KY780931.1 | China / Китай | 2016 | MN654379.1 | South Korea / Южная Корея | 2009 |
KY780932.1 | China / Китай | 2016 | KX494979.1 | South Korea / Южная Корея | 2016 |
KY780933.1 | China / Китай | 2016 | KY471318.1 | South Korea / Южная Корея | 2017 |
PP002039.1 | China / Китай | 2018 | KY471322.1 | South Korea / Южная Корея | 2017 |
PP002041.1 | China / Китай | 2018 | KY471319.1 | South Korea / Южная Корея | 2017 |
MN052861.1 | China / Китай | 2018 | KY471320.1 | South Korea / Южная Корея | 2017 |
MK123978.1 | China / Китай | 2018 | KY471321.1 | South Korea / Южная Корея | 2017 |
PP002033.1 | China / Китай | 2018 | KY471323.1 | South Korea / Южная Корея | 2017 |
PP002034.1 | China / Китай | 2018 | MW053454.1 | South Korea / Южная Корея | 2019 |
PP002038.1 | China / Китай | 2018 | MN654393.1 | Japan / Япония | 2012 |
PP002032.1 | China / Китай | 2018 |
Results
In a sample of respiratory specimens received from hospitals in Moscow during the 2021–2022 season, 12 contained HAdV according to qPCR data. The samples were analyzed for viral load values and the multiplicity of infection. One of the samples (bronchoalveolar lavage fluid from a patient hospitalized with pneumonia), characterized by a high level of HAdV DNA load (cycle threshold value, Ct = 12.3) and the absence of co-infection with other respiratory viruses, was used for HAdV isolation.
The isolated adenovirus was identified as HAdV-55 based on sequencing data, deposited in the State Collection of Viruses at the D.I. Ivanovsky Institute of Virology of the N.F. Gamaleya NRCEM of the Ministry of Health of Russia under the number SCV3008:Ad55, and the genomic data were registered in GenBank (registration number PQ641625).
RFLP analysis of the isolate DNA revealed the following fragments: Cfr41I (12894, 8674, 7768, 2664, 1194, 1181 and 403 bp), XhoI (10335, 8005, 6544, 5761, 2628, 1355, and 150 bp) and XagI (20615, 5219, 3789, 2944, 1359 and 852 bp). Fig. 1 (a) presents the most prominent of them. Comparison with in silico data for isolates HAdV-55 (MG905110), HAdV-11 (AY163756) and HAdV-14 (MF062484) (Fig. 1 b) confirms the similarity of the restriction fragments of isolates SCV3008:Ad55 and HAdV-55 (MG905110) and emphasizes that the genome of the recombinant HAdV-55 is primarily composed of HAdV-14 genes.
Fig. 1. Restriction fragment length polymorphisms (RFLPs) using Cfr41I, XagI, and XhoI restriction enzymes. a – DNA of the SCV3008 isolate in vitro; b – DNA of the HAd-55, HAd-11 and HAd-14 strains in silico.
Рис. 1. Полиморфизм длин рестрикционных фрагментов, полученных с рестриктазами Cfr41I, XagI и XhoI. a ‒ для ДНК изолята SCV3008:Ad55 in vitro; б ‒ для ДНК штаммов HAd-55, HAd-11 и HAd-14 in silico.
The similarity with HAdV-14 was also demonstrated by the ANI calculation between the genomes of the HAdV-55 samples presented in Table 1, SCV3008:Ad55, and HAdV-14 (MF062484). The ANI value between HAdV-55 and HAdV-14 was 98.7478–98.9411, and between the genomes of HAdV-55 it was 99.6546–100.000.
The phylogenetic analysis included HAdV-55 isolates collected in 8 countries from 1969 to 2022 (Table 1). The phylogenetic tree is illustrated in Fig. 2, which shows that the genomes formed 4 clades. The ancestral clade “0” included the earliest isolates MG905110 (Spain, 1969), MN654394 (USA, 1976), and the isolate MN654395 (USA, 2020). Clade I was formed by 13 isolates from South Korea and two isolates from China (2011 and 2015). Clade II, which was primarily composed of isolates from Egypt (12 isolates), was the most diverse in terms of country representation. It included isolates from Singapore (MN654388 and MN654389, 2005), Japan (MN654393, 2012), the USA (MN654392, 1997; MT513753, 2006), China (JX123027, 2010), and the isolate SCV3008:Ad55 identified by us. The most numerous clade III was formed by isolates from China from 2006 to 2021.
Fig. 2. Neighbor-joining phylogenetic tree constructed based on the complete genomes of 83 HAd-55 isolates presented in Table 1, and the genome of the SCV3008:Ad55 isolate (PQ641625). Clades 0–III are characterized in the legend. MF062484 – HAd-14 isolate, represents an outgroup.
Рис. 2. Филогенетическое древо Neighbor-joining, построенное на основе полных геномов 83 изолятов HAd-55, представленных в табл. 1, и генома изолята SCV3008:Ad55 (PQ641625). Клады 0–III охарактеризованы в легенде. MF062484 – изолят HAd-14, представляет внешнюю группу.
In clade II, the isolate closest to SCV3008:Ad55 in terms of ANI (99.9396) was the isolate from Japan (MN654393). Differences between the genomes SCV3008:Ad55 and MN654393 were identified both in the untranslated regions of the genome and in the genes of structural and non-structural proteins (Table 2). The identified substitutions were compared with the sequences of other genomes in the sample. Note that the comparison of the ITR regions and the nearest ones was not possible for all genomes. Of the identified substitutions, 4 were characteristic of most isolates of clade II, 4 were unique to the genome of the Japanese isolate, and 12 substitutions distinguished the SCV3008:Ad55 isolate.
Table 2. Characterization of substitutions in the genome of isolate SVC308-Ad55
Таблица 2. Характеристика замен в геноме изолята SVC308:Ad55
Name of the region of the genome/CDS Название области генома/CDS | Substitution in the SVC308:Ad55 genome relative to the WPAFB415 Замена в геноме SVC308:Ad55 относительно генома WPAFB415 | Mutation frequency Встречаемость мутации | Substitutions in amino acid sequence Замены в последовательности белка |
Untranslated regions Нетранслируемые области | C134T** | Clade Egypt* | |
A445G | SVC308:Ad55 | ||
T3437C | SVC308:Ad55 | ||
A3924G | SVC308:Ad55 | ||
G34619T*** | SVC308:Ad55 | ||
pIX 14.2 kDa / 14,2 кДа | C3536T | SVC308:Ad55 | |
pIVa2 50.9 kDa / 50,9 кДа | T4656C | WPAFB415 | |
128,9 кДа ДНК-полимераза 128.9 kDa DNA polymerase | C6707T | SVC308:Ad55 | |
T6764A | WPAFB415 | Lys→Asp | |
pTP 73.4 kDa / 73,4 кДа | G8779A | SVC308:Ad55 | |
C8815T | Clade Egypt* | ||
L1 52/55K 43,9 kDa / 43,9 кДа | C11734T | SVC308:Ad55 | |
L1 pIIIa 65.6 kDa / 65,6 кДа | G13349A | Clade Egypt* | |
L2 62,5 kDa penton protein / пентон 62,5 кДа | T13787A | SVC308:Ad55 | Thr→Ala |
G14203A | SVC308:Ad55 | ||
L2 pV 40.1 kDa / 40,1 кДа | T16315C | WPAFB415 | Glu→Leu |
G16530C | WPAFB415 | ||
58.3 kDa DNA-binding protein E2A 58,3 кДа ДНК-связывающий белок | G22776A | SVC308:Ad55 | |
L4 22K 21.6 kDa / 21,6 кДа | C26083T | Clade Egypt* | His→Tyr |
L4 pVIII 25 kDa / 25 кДа | A26918G | WPAFB415 | |
E3 18.5 kDa / 18,5 kDa | T28096G | SVC308:Ad55 |
Note. * – except for MT513753, MN654393, JX123027; ** – the left ITR region is found in only 65 of the 85 genomes; *** – the region of the genome in front of the right ITR is present in only 75 out of 85 genomes.
Примечание. * – кроме MT513753, MN654393, JX123027; ** – область левого ITR есть только в 65 геномах из 85; *** – область генома перед правым ITR присутствует только в 75 геномах из 85.
The substitutions in 4 reading frames were non-synonymous. The mutation in the peptide (Thr29Ala) was unique to the isolate SCV3008:Ad55. Changes in the DNA polymerase (Asp566Lys) and in the pV protein (Leu105Glu) distinguished the Japanese isolate MN654393. The mutation in the L4 22K protein (His162Tyr) was found in 14 isolates of clade II, including SCV3008:Ad55 (Table 2).
In the analysis of genomes, we noticed the heterogeneity in the sizes of poly-A/poly-T sequences in intergenic regions (Table 3). The sizes of regions 2–6, marked in the SCV3008:Ad55 genome, were characteristic of many genomes in the HAdV-55 sample. Region 1 with the A6G substitution was unique to SCV3008:Ad55. Homopolymer sequences in adenovirus genomes, also known as microsatellites, drew the attention of researchers during the investigation of adenovirus infection outbreaks with fatal outcomes in U.S. military cohorts in 2006–2007. Polymorphism of microsatellite loci lengths became a high-resolution marker for attributing HAdV-14 to a single outbreak [18]. A comparison of microsatellite loci in the isolates of clade II «Egypt» was conducted, which included 17 isolates from different continents. The data in Table 4 indicate that most isolates of clade II were similar in microsatellite size across all 6 loci. The maximum number of loci (4) distinguished the genomes of isolates from Japan (MN654393) and Russia (SCV3008:Ad55), the genome of the single isolate from China in clade II (JX123027) differed by 3 loci, the isolates from the USA (MN654392, 1997; MT513753, 2006) differed by 2 different loci, and the isolates from Singapore (MN654388 and MN654389) differed by 1 locus each. Of the 10 Egyptian isolates in clade II, three had one locus difference each. Thus, with the high conservatism of HAdV-55 genomes, microsatellite loci indeed allow for the differentiation of virus genomes within a single clade.
Table 3. Regions of repeats in the genomes of a sample of adenoviruses belonging to genotype 55
Таблица 3. Области повторов в геномах выборки аденовирусов человека 55-го типа
N | The position according to PQ641625 genome / Neighboring ORS Положение по геному PQ641625 / Соседние ОРС | Isolate Изолят | The number of nucleotides in the repeat Число нуклеотидов в повторе | Number of isolates Количество изолятов |
1 | 3918–3933 bp / pIX; pIVa2 | MF062484/China/2010* | 1 | |
OP375144/China/2021 | 3 | |||
MW053454/China/2023 | 1 | |||
PQ641625/Russia/2022 | 1 | |||
MN654393/Japan/2012 | 1 | |||
MN654378/S.Korea/2009 | 24 | |||
MN654392/USA/1997 | 5 | |||
MN654394/USA/1976 | 1 | |||
FJ643676/China/2011 | 48 | |||
2 | 10651–10664 bp / pTP; L1 | MF062484/China/2010* | ||
MN654395/USA/2020 | 1 | |||
MW053454/China/2023 | 3 | |||
KY471322/S.Korea/2017 | 9 | |||
PP002032/China/2018 | 21 | |||
PQ641625/Russia/2022 | 7 | |||
MN654394/USA/1976 | 37 | |||
MK123980/China/2013 | 6 | |||
KP896484/China/2013 | 1 | |||
3 | 13620–13630 bp/L1 pIIIa; L2 penton [polyA_signal_sequence (aaataaa) 13627–13633 bp] | MF062484/China/2010* | 1 | |
PP002034/China/2018 | 1 | |||
MN654388/Singapore/2005 | 16 | |||
PQ641625/Russia/2022 | 66 | |||
PP002040/China/2018 | 1 | |||
4a | 17323–17334 bp / L2 pX; L3 pVI | MF062484/China/2010* | ||
MK123978/China/2018 | 1 | |||
PQ641625/Russia/2022 | 51 | |||
KY070248/China/2016 | 20 | |||
MN654394/USA/1976 | 12 | |||
KP896484/China/2013 | 1 | |||
4b | 17341–17352 bp / L2 pX; L3 pVI [polyA_signal_sequence (aataaa) 17339–17344 bp] | MF062484/China/2010* | ||
KX494979/S.Korea/2016 | 5 | |||
PQ641625/Russia/2022 | 75 | |||
KH289874/China/2015 | 5 | |||
5 | 29474–29486 bp / E3 20.2 kDa; E3 10.3 kDa | MF062484/China/2010* | ||
MT513753/USA/2006 | 1 | |||
MK123981/China/2013 | 3 | |||
OM714808/China/2020 | 1 | |||
PQ641625/Russia/2022 | 17 | |||
KP896483/China/2013 | 37 | |||
JX123029/China/2012 | 25 | |||
KP896484/China/2013 | 1 | |||
6 | 34006 – 34016 bp / E4 ORF2; E4 ORF1 | MF062484/China/2010* | 1 | |
MT513753/USA/2006 | 3 | |||
PQ641625/Russia/2022 | 28 | |||
JX123029/China/2012 | 53 |
Note. * – the ancestral genome of human adenovirus 14.
Примечание. * – предковый геном Human adenovirus 14.
Table 4. Size of microsatellite loci in the genomes of clade II «Egypt» isolates
Таблица 4. Размер локусов микросателлитов в геномах изолятов клады II «Egypt»
Locus Локус | Isolate Изолят | Homopolymer size (nt) Размер гомополимера (нт) | Locus Локус | Isolate Изолят | Homopolymer size (nt) Размер гомополимера (нт) |
1 | Most / Большинство* | A (13) | 4a, 4b | Most / Большинство* | A (10); A (10) |
MN654380 (Egypt 2000) | A (12) | JX123027 (China 2010) | A (10); A (11) | ||
MN654392 (USA 1997) | A (12) | MN654392 (USA 1997) | A (9); A (10) | ||
MN654393 (Japan 2012) | A (14) | 5 | Большинство / Most | T (9) | |
PQ641625 (Russia 2022) | A (13) G (1) | MN654393 (Japan 2012) | T (11) | ||
2 | Most / Большинство* | T (11) | PQ641625 (Russia 2022) | T (11) | |
MN654393 (Japan 2012) | T (12) | JX123027 (China 2010) | T (10) | ||
PQ641625 (Russia 2022) | T (12) | MT513753 (USA 2006) | T (14) | ||
JX123027 (China 2010) | T (12) | 6 | Большинство / Most | A (8) | |
3 | Большинство / Most | A (10) | MN654393 (Japan 2012) | A (10) | |
MN654386 (Egypt 2007) | A (11) | PQ641625 (Russia 2022) | A (10) | ||
MN654387 (Egypt 2009) | A (11) | MT513753 (USA 2006) | A (11) | ||
MN654388 (Singapore 2005) | A (11) | ||||
MN654389 (Singapore 2005) | A (11) |
Note. * ‒ Clade II «Egypt» contains 17 isolates.
Примечание. * ‒ в кладе II «Egypt» 17 изолятов.
Discussion
For the first time, the presented study describes the genome of the HAdV-55 SCV3008:Ad55 strain isolated in the territory of the Russian Federation. It should be noted that previous molecular-epidemiological genomic studies of adenoviruses in the Russian Federation are rare and focused on the study of M. caesari HAdV, pathogens of respiratory infections in children [19]. The collection of comparative information for genomic studies is hindered by the low level of implementation of genotyping methods in the laboratory diagnosis of adenovirus infection.
In Russia, the molecular-genetic approach, approved since 2010, is used for epidemiological monitoring for adenovirus infection and identification of the pathogen up to the family Adenoviridae at the Reference Center for Influenza and ARVI Diagnosis based at the A.A. Smorodintsev Research Institute of Influenza, at the Center for Ecology and Epidemiology of Influenza at the N.F. Gamaleya National Research Center for Epidemiology and Microbiology, and at the reference bases of Rospotrebnadzor. Adenovirus infections in children are under special control and are also subject to molecular diagnostics according to clinical guidelines2. However, HAdV genotyping is not included in the list of laboratory diagnostic methods.
ECDC (European Centre for Disease Prevention and Control) does not conduct routine surveillance of adenovirus infections and only records outbreaks of the disease, whereas the CDC (Centers for Disease Control and Prevention, USA) has developed guidelines for identifying HAdV based on nucleic acid amplification and has created the National Adenovirus Type Reporting System (NATRS). According to NATRS data from 2017 to 2023, HAdV 6 genotypes were the most common in the USA, among which HAdV-7 and HAdV-14 of the M. blackbeardi species accounted for 13.4% and 7.8%, respectively (https://www.cdc.gov/adenovirus/hcp/outbreaks/index.html). The Japanese National Infectious Disease Epidemiological Surveillance System also conducts adenovirus genotyping, noting among the predominant M. blackbeardi HAdV-3, 7, 11, 34, 35, and among the minor ones 14, 16, 55, 66, 68, 79 [20]. The CDC of China monitors influenza and ARVI, but does not publish reports on virus genotyping in the public domain [21]. Thus, among national surveillance systems, only Japan’s epidemiological surveillance system monitors HAdV-55.
Analysis of scientific publications from 2012 to 2025, available on PubMed, showed that out of 48 articles mentioning HAdV-55 in their keywords, 39 (81%) were published by researchers from China, 7 from South Korea, and one each from the USA and Senegal. This ratio of publications confirms the endemicity of HAdV-55 in China and South Korea. It should be noted that among the publications from South Korea, only two present a study of HAdV-55 infections among the civilian population, while the others describe outbreaks of ARVI caused by HAdV-55 among military personnel [22]. The topic of HAdV-55 infections among military personnel is continued by a publication from the USA [23], dedicated to the analysis of the MW053454 virus isolate, which was isolated from an American serviceman who was in South Korea in 2019. The MW053454 isolate differed from the South Korean isolate KX494979 from 2016 by one synonymous substitution. In the current study, both isolates were placed in clade I «South Korea». In Senegal, from 2012 to 2015, M. blackbeardi HAdV was identified in 9 cases of patients with ARVI, among which HAdV-7, HAdV-55 and HAdV-11 were noted [24]. The presented data indicate that HAdV genotyping is gradually being integrated into laboratory practice.
Considering the above, in order to conduct a comparative study of the genome of the isolate SCV3008:Ad55, GenBank data was used, collecting a sample of 83 isolates from 1969 to 2022 from 7 countries. The analysis of the sample showed a high similarity of HAdV-55 genomes, reaching 99.7–100% on the ANI scale, which is consistent with data from other studies conducted on a smaller number of isolates [25]. At the same time, the phylogenetic analysis allowed for the division of the genomes in the sample into clades, indicating the presence of heterogeneity even with high homology. Clades I and III corresponded to the geographical origin of the isolates and showed an epidemiological connection both between the isolates from China and those from South Korea. Isolates from Egypt from 2000 to 2009, a country distant from areas endemic to HAdV-55, clustered with isolates from five countries, including China, indicating the spread of HAdV-55, facilitated by globalization processes. It should be noted that the isolates in cluster II predominantly came from the civilian population. The exceptions were the isolates from Singapore and Japan, obtained from samples of military personnel who fell ill with ARVI [25].
Comparative genomic analysis revealed differences in the SVC3008:Ad55 isolate, specifically 12 point mutations distributed throughout the genome, of which a non-synonymous substitution occurred in the reading frame of the 62.5 kDa penton L2, leading to a T29A substitution at the N-terminus of the protein sequence. Since the substitution is conservative, it does not affect the amphipathic properties of the N-terminal helix of the protein or the capability of the PPRY motif (42–45 a.a.) to interact with the WW domains of cellular ubiquitin ligases, which facilitates the virus’s entry into the eukaryotic cell, determining its infectivity [26].
Additional information about the diversity of closely related genomes of the clade II isolates was provided by the analysis of microsatellite length polymorphism (homopolymers) at 6 loci in the untranslated regions. Out of 17 isolates of the clade, 10 had differences in at least one locus. The isolates from Japan and Russia differed from the other isolates of the clade by 4 microsatellite loci. This approach even allowed for the differentiation of isolates from Egypt within two regions: Alexandria (2000–2002) and Cairo (2005–2009).
Conclusion
Comparative genomic study of HAdV-55 isolates, which emerged as a result of the recombination of HAdV-14 and HAdV-11, showed genome stability since 1969 and a slow accumulation of mutations in both coding and non-coding regions, allowing the identification of unique substitutions in the new SVC3008:Ad55 isolate. The obtained genomic information laid the foundation for the development of diagnostic kits and further monitoring for HAdV-55, which causes infections complicated by bronchopneumonia. At the same time, since adenoviruses are subject to recombinational variability and there are multiple recombination hotspots (genes of the penton, hexon, fiber, E1, E3 and E4) [27], whole-genome sequencing is particularly effective in monitoring and molecular epidemiological analysis of adenovirus pathogens.
1 ICTV. Family: Adenoviridae. Available at: https://ictv.global/report/chapter/adenoviridae/adenoviridae
2 Clinical guidelines (treatment protocol) for providing medical care to children with adenoviral infection; 2013. Available at: http://niidi.ru/dotAsset/69f7f879-9765-4634-a621-8792acf587b7.pdf
About the authors
Daniil A. Shein
National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya of the Ministry of Health of the Russian Federation
Email: daniil.schein@yandex.ru
ORCID iD: 0009-0003-3768-9817
Postgraduate Student, Laboratory of genome analysis
Russian Federation, Moscow, 123098Natalya N. Ryzhova
National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya of the Ministry of Health of the Russian Federation
Email: rynatalia@yandex.ru
ORCID iD: 0000-0001-5361-870X
Cand. Sci. (Biol.), Senior Researcher, Laboratory of genome analysis
Russian Federation, Moscow, 123098Marina S. Kunda
National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya of the Ministry of Health of the Russian Federation
Email: markunda99@gmail.com
ORCID iD: 0000-0003-1945-0397
Cand. Sci. (Biol.), Senior Researcher, Laboratory of genome analysis
Russian Federation, Moscow, 123098Ekaterina I. Ermolova
National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya of the Ministry of Health of the Russian Federation
Email: aksenova16@yandex.ru
ORCID iD: 0000-0002-0437-9404
Cand. Sci. (Biol.), Senior Researcher, Laboratory of genome analysis
Russian Federation, Moscow, 123098Tatiana A. Ozharovskaia
National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya of the Ministry of Health of the Russian Federation
Email: t.ozh@yandex.ru
ORCID iD: 0000-0001-7147-1553
Cand. Sci. (Biol.), Senior Researcher, Immunobiotechnology laboratory
Russian Federation, Moscow, 123098Olga Popova
National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya of the Ministry of Health of the Russian Federation
Email: olga.popova31@yandex.ru
ORCID iD: 0000-0003-3248-1227
junior researcher, Immunobiotechnology laboratory
Russian Federation, Moscow, 123098Natalia A. Nikitenko
National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya of the Ministry of Health of the Russian Federation
Email: nan-nikitenko@yandex.ru
ORCID iD: 0000-0001-5829-744X
Cand. Sci. (Biol.), Senior Researcher, Head of Medical Department
Russian Federation, Moscow, 123098Kirill G. Krasnoslobodtsev
National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya of the Ministry of Health of the Russian Federation
Email: kkg_87@mail.ru
ORCID iD: 0000-0003-1745-9128
researcher, Influenza etiology and epidemiology laboratory
Russian Federation, Moscow, 123098Elena I. Burtseva
National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya of the Ministry of Health of the Russian Federation
Email: elena-burtseva@yandex.ru
ORCID iD: 0000-0003-2518-6801
Dr. Sci. (Medicine), Head, Influenza etiology and epidemiology laboratory
Russian Federation, Moscow, 123098Olga V. Zubkova
National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya of the Ministry of Health of the Russian Federation
Author for correspondence.
Email: olga-zubkova@yandex.ru
ORCID iD: 0000-0001-7893-8419
Cand. Sci. (Biol.), Leading Researcher, Immunobiotechnology laboratory
Russian Federation, Moscow, 123098Olga L. Voronina
National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya of the Ministry of Health of the Russian Federation
Email: olv550@gmail.com
ORCID iD: 0000-0001-7206-3594
Cand. Sci. (Biol.), Assistant Professor, Leading Researcher, Head of Laboratory of genome analysis
Russian Federation, Moscow, 123098Alexander L. Gintsburg
National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya of the Ministry of Health of the Russian Federation
Email: gintsburg@gamaleya.org
ORCID iD: 0000-0003-1769-5059
Dr. Sci. (Biol.), RAS academician, professor, Director
Russian Federation, Moscow, 123098References
- Lynch J.P. 3rd, Kajon A.E. Adenovirus: epidemiology, global spread of novel serotypes, and advances in treatment and prevention. Semin. Respir. Crit. Care Med. 2016; 37(4): 586–602. https://doi.org/10.1055/s-0036-1584923
- Coleman K.K., Wong C.C., Jayakumar J., Nguyen T.T., Wong A.W.L., Yadana S., et al. Adenoviral infections in Singapore: Should new antiviral therapies and vaccines be adopted? J. Infect. Dis. 2020; 221(4): 566–77. https://doi.org/10.1093/infdis/jiz489
- Xu W., Xu Z., Huang L., Qin E.Q., Zhang J.L., Zhao P., et al. Transcriptome sequencing identifies novel immune response genes highly related to the severity of human adenovirus type 55 infection. Front. Microbiol. 2019; 10: 130. https://doi.org/10.3389/fmicb.2019.00130
- Kajon A.E., Lamson D.M., St. George K. Emergence and re-emergence of respiratory adenoviruses in the United States. Curr. Opin. Virol. 2019; 34: 63–9. https://doi.org/10.1016/j.coviro.2018.12.004
- Dhingra A., Hage E., Ganzenmueller T., Böttcher S., Hofmann J., Hamprecht K., et al. Molecular Evolution of Human Adenovirus (HAdV) Species C. Sci Rep. 2019; 9(1): 1039. https://doi.org/10.1038/s41598-018-37249-4
- Scott M.K., Chommanard C., Lu X., Appelgate D., Grenz L., Schneider E., et al. Human adenovirus associated with severe respiratory infection, Oregon, USA, 2013–2014. Emerg. Infect. Dis. 2016; 22(6): 1044–51. https://doi.org/10.3201/eid2206.151898
- Hierholzer J.C., Pumarola A., Rodriguez-Torres A., Beltran M. Occurrence of respiratory illness due to an atypical strain of adenovirus type 11 during a large outbreak in Spanish military recruits. Am. J. Epidemiol. 1974; 99(6): 434–42. https://doi.org/10.1093/oxfordjournals.aje.a121632
- Li Q.G., Hambraeus J., Wadell G. Genetic relationship between thirteen genome types of adenovirus 11, 34, and 35 with different tropisms. Intervirology. 1991; 32(6): 338–50. https://doi.org/10.1159/000150218
- Yang Z., Zhu Z., Tang L., Wang L., Tan X., Yu P., et al. Genomic analyses of recombinant adenovirus type 11a in China. J. Clin. Microbiol. 2009; 47(10): 3082–90. https://doi.org/10.1128/JCM.00282-09
- Seto D., Chodosh J., Brister J.R., Jones M.S. Using the whole-genome sequence to characterize and name human adenoviruses. J. Virol. 2011; 85(11): 5701–2. https://doi.org/10.1128/JVI.00354-11
- Seto D., Jones M.S., Dyer D.W., Chodosh J. Characterizing, typing, and naming human adenovirus type 55 in the era of whole genome data. J. Clin. Virol. 2013; 58(4): 741–2. https://doi.org/10.1016/j.jcv.2013.09.025
- Centers for Disease Control and Prevention (CDC). Civilian outbreak of adenovirus acute respiratory disease – South Dakota, 1997. MMWR Morb. Mortal. Wkly Rep. 1998; 47(27): 567–70.
- Kajon A.E., Mistchenko A.S., Videla C., Hortal M., Wadell G., Avendaño L.F. Molecular epidemiology of adenovirus acute lower respiratory infections of children in the south cone of South America (1991–1994). J. Med. Virol. 1996; 48(2): 151–6. https://doi.org/10.1002/(sici)1096-9071(199602)48:2%3C151::aid-jmv6%3E3.0.co;2-8
- Salama M., Amitai Z., Nutman A., Gottesman-Yekutieli T., Sherbany H., Drori Y., et al. Outbreak of adenovirus type 55 infection in Israel. J. Clin. Virol. 2016; 78: 31–5. https://doi.org/10.1016/j.jcv.2016.03.002
- Jing S., Zhang J., Cao M., Liu M., Yan Y., Zhao S., et al. Household transmission of human adenovirus type 55 in case of fatal acute respiratory disease. Emerg. Infect. Dis. 2019; 25(9): 1756–8. https://doi.org/10.3201/eid2509.181937
- Burtseva E.I., Panova A.D., Kolobukhina L.V., Ignatjeva A.V., Kirillova E.S., Breslav N.V., et al. Epidemic season 2021–2022: Frequency of co-infection by respiratory viral pathogens. Epidemiologiya i infektsionnye bolezni. 2023; 28(2): 67–77. https://doi.org/10.17816/EID321873 (in Russian)
- Tamura K., Stecher G., Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021; 38(7): 3022–7. https://doi.org/10.1093/molbev/msab120
- Houng H.S., Lott L., Gong H., Kuschner R.A., Lynch J.A., Metzgar D. Adenovirus microsatellite reveals dynamics of transmission during a recent epidemic of human adenovirus serotype 14 infection. J. Clin. Microbiol. 2009; 47(7): 2243–8. https://doi.org/10.1128/JCM.01659-08
- Kurskaya O.G., Prokopyeva E.A., Dubovitskiy N.A., Solomatina M.V., Sobolev I.A., Derko A.A., et al. Genetic Diversity of the Human Adenovirus C Isolated from Hospitalized Children in Russia (2019-2022). Viruses. 2024; 16(3): 386. https://doi.org/10.3390/v16030386
- Adenovirus infections, 2008 to 2020, Japan. IASR. 2021; 42(4): 67–9. Available at: https://id-info.jihs.go.jp/niid/en/iasr/12459-494te.html
- Sun H., Hu W., Wei Y., Hao Y. Review: Drawing on the development experiences of infectious disease surveillance systems around the world. China CDC Wkly. 2024; 6(41): 1065–74. https://doi.org/10.46234/ccdcw2024.220
- Ko J.H., Woo H.T., Oh H.S., Moon S.M., Choi J.Y., Lim J.U., et al. Ongoing outbreak of human adenovirus-associated acute respiratory illness in the Republic of Korea military, 2013 to 2018. Korean J. Intern. Med. 2021; 36(1): 205–13. https://doi.org/10.3904/kjim.2019.092
- Hughes J.J., Yang Y., Fries A.C., Maljkovic Berry I., Pollio A.R., Fung C.K., et al. Complete genome sequences of two human adenovirus type 55 isolates from South Korea and the United States. Microbiol. Resour. Announc. 2021; 10(5): e01347-20. https://doi.org/10.1128/MRA.01347-20
- Niang M.N., Diop N.S., Fall A., Kiori D.E., Sarr F.D., Sy S., et al. Respiratory viruses in patients with influenza-like illness in Senegal: Focus on human respiratory adenoviruses. PLoS One. 2017; 12(3): e0174287. https://doi.org/10.1371/journal.pone.0174287
- Hang J., Kajon A.E., Graf P.C.F., Berry I.M., Yang Y., Sanborn M.A., et al. Human adenovirus type 55 distribution, regional persistence, and genetic variability. Emerg. Infect. Dis. 2020; 26(7): 1497–505. https://doi.org/10.3201/eid2607.191707
- Wodrich H., Henaff D., Jammart B., Segura-Morales C., Seelmeir S., Coux O., et al. A capsid-encoded PPxY-motif facilitates adenovirus entry. PLoS Pathog. 2010; 6(3): e1000808. https://doi.org/10.1371/journal.ppat.1000808
- Wang F., De R., Han Z., Xu Y., Zhu R., Sun Y., et al. High-frequency recombination of human adenovirus in children with acute respiratory tract infections in Beijing, China. Viruses. 2024; 16(6): 828. https://doi.org/10.3390/v16060828
Supplementary files