Vol 66, No 6 (2021)
- Year: 2021
- Published: 09.12.2021
- Articles: 8
- URL: https://virusjour.crie.ru/jour/issue/view/53
Full Issue
REVIEWS
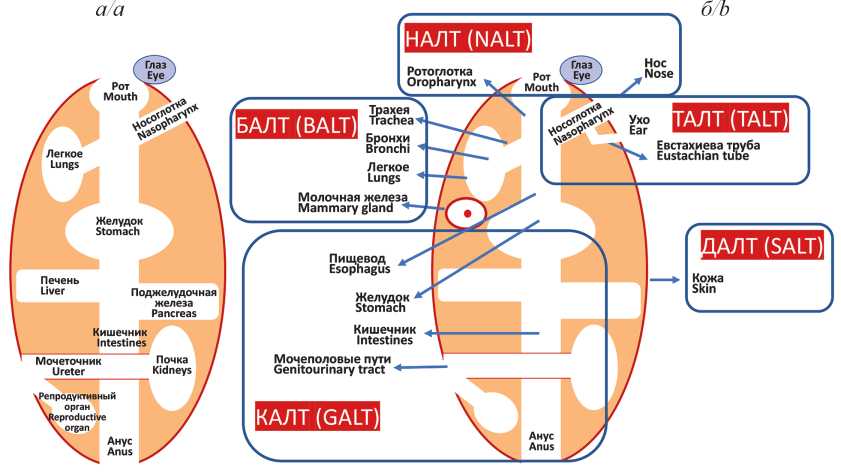
Mucosal immunity and vaccines against viral infections
Abstract
Mucosal immunity is realized through a structural and functional system called mucose-associated lymphoid tissue (MALT). MALT is subdivided into parts (clusters) depending on their anatomical location, but they all have a similar structure: mucus layer, epithelial tissue, lamina propria and lymphoid follicles. Plasma cells of MALT produce a unique type of immunoglobulins, IgA, which have the ability to polymerize. In mucosal immunization, the predominant form of IgA is a secretory dimer, sIgA, which is concentrated in large quantities in the mucosa. Mucosal IgA acts as a first line of defense and neutralizes viruses efficiently at the portal of entry, preventing infection of epithelial cells and generalization of infection. To date, several mucosal antiviral vaccines have been licensed, which include attenuated strains of the corresponding viruses: poliomyelitis, influenza, and rotavirus. Despite the tremendous success of these vaccines, in particular, in the eradication of poliomyelitis, significant disadvantages of using attenuated viral strains in their composition are the risk of reactogenicity and the possibility of reversion to a virulent strain during vaccination. Nevertheless, it is mucosal vaccination, which mimics a natural infection, is able to induce a fast and effective immune response and thus help prevent and possibly stop outbreaks of many viral infections. Currently, a number of intranasal vaccines based on a new vector approach are successfully undergoing clinical trials. In these vaccines, the safe viral vectors are used to deliver protectively significant immunogens of pathogenic viruses. The most tested vector for intranasal vaccines is adenovirus, and the most significant immunogen is SARSCoV-2 S protein. Mucosal vector vaccines against human respiratory syncytial virus and human immunodeficiency virus type 1 based on Sendai virus, which is able to replicate asymptomatically in cells of bronchial epithelium, are also being investigated.
 399-408
399-408


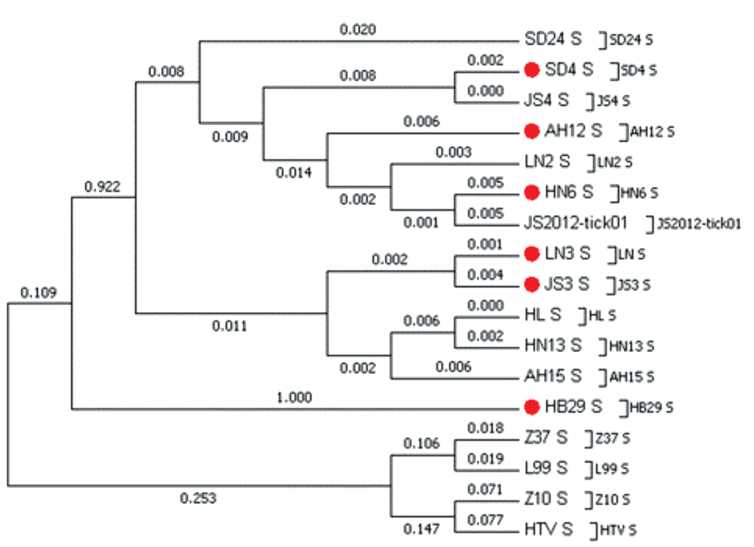
The molecular evolution of Dabie bandavirus (Phenuiviridae: Bandavirus: Dabie bandavirus), the agent of severe fever with thrombocytopenia syndrome
Abstract
Since the Dabie bandavirus (DBV; former SFTS virus, SFTSV) was identified, the epidemics of severe fever with thrombocytopenic syndrome (SFTS) caused by this virus have occurred in several countries in East Asia. The rapid increase in incidence indicates that this infectious agent has a pandemic potential and poses an imminent global public health threat.
The analysis of molecular evolution of SFTS agent that includes its variants isolated in China, Japan and South Korea was performed in this review. The evolution rate of DBV and the estimated dates of existence of the common ancestor were ascertained, and the possibility of reassortation was demonstrated.
The evolutionary rates of DBV genome segments were estimated to be 2.28 × 10-4 nucleotides/site/year for S-segment, 2.42 × 10-4 for M-segment, and 1.19 × 10-4 for L-segment. The positions of positive selection were detected in the viral genome.
Phylogenetic analyses showed that virus may be divided into two clades, containing six different genotypes. The structures of phylogenetic trees for S-, M- and L-segments showed that all genotypes originate from the common ancestor.
Data of sequence analysis suggest that DBV use several mechanisms to maintain the high level of its genetic diversity. Understanding the phylogenetic factors that determine the virus transmission is important for assessing the epidemiological characteristics of the disease and predicting its possible outbreaks.
 409-416
409-416


ORIGINAL RESEARCHES
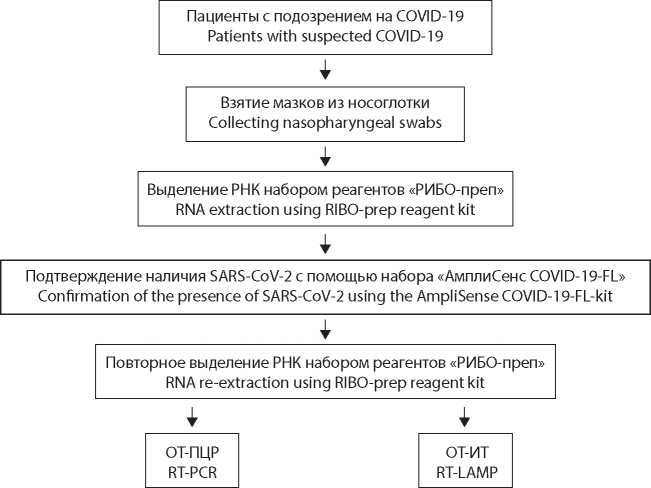
Molecular methods for diagnosing novel coronavirus infection: comparison of loop-mediated isothermal amplification and polymerase chain reaction
Abstract
Introduction. Currently, the basis for molecular diagnostics of most infections is the use of reverse transcription polymerase chain reaction (RT-PCR). Technologies based on reverse transcription isothermal loop amplification (RT-LAMP) can be used as an alternative to RT-PCR for diagnostic purposes. In this study, we compared the RTLAMP and RT-PCR methods in order to analyze both the advantages and disadvantages of the two approaches.
Material and methods. For the study, we used reagent kits based on RT-PCR and RT-LAMP. The biological material obtained by taking swabs from the mucous membrane of the oropharynx and nasopharynx in patients with symptoms of a new coronavirus infection was used.
Results. We tested 381 RNA samples of the SARS-CoV-2 virus (Coronaviridae: Coronavirinae: Betacoronavirus; Sarbecovirus) from various patients. The obtained values of the threshold cycle (Ct) for RT-PCR averaged 20.0 ± 3.7 s (1530 ± 300 s), and for RT-LAMP 12.8 ± 3.7 s (550 ± 160 s). Proceeding from the theoretical assumptions, a linear relationship between values obtained in two kits was proposed as a hypothesis; the correlation coefficient was approximately 0.827. At the same time, for samples with a low viral load (VL), the higher Ct values in RT-LAMP did not always correlated with those obtained in RT-PCR.
Discussion. We noted a significant gain in time for analysis using RT-LAMP compared to RT-PCR, which can be important in the context of testing a large number of samples. Being easy to use and boasting short turnaround time, RT-LAMP-based test systems can be used for mass screening in order to identify persons with medium and high VLs who pose the greatest threat of the spread of SARS-CoV-2, while RT-PCR-based diagnostic methods are also suitable for estimation of VL and its dynamics in patients with COVID-19.
 417-424
417-424


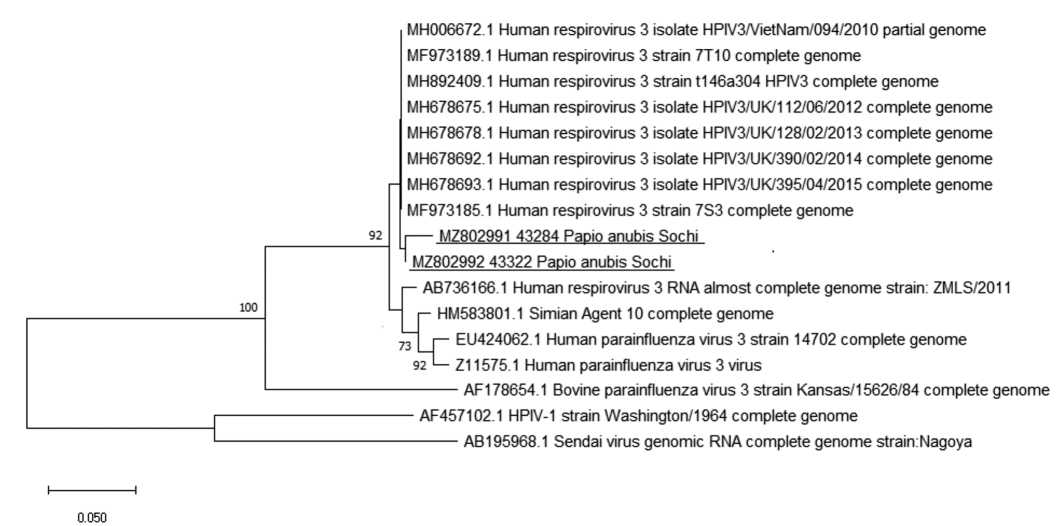
Prevalence of laboratory markers of human respiratory viruses in monkeys of Adler primate center
Abstract
Introduction. The relevance of studying the circulation of human respiratory viruses among laboratory primates is associated with the need to test vaccines and antiviral drugs against these infections on monkeys.
The aim of this work was to study the prevalence of serological and molecular markers of human respiratory viral infections in laboratory primates born at the Adler Primate Center and in imported monkeys.
Material and methods. Blood serum samples (n = 1971) and lung autopsy material (n = 26) were obtained from different monkey species. These samples were tested for the presence of serological markers of measles, parainfluenza (PI) types 1, 2, 3, influenza A and B, respiratory syncytial (RS) and adenovirus infections using enzyme immunoassay (ELISA). Detection of RS virus, metapneumovirus, PI virus types 1–4, rhinovirus, coronavirus, and adenoviruses B, C, E and bocavirus nucleic acids in this material was performed by reverse transcription polymerase chain reaction (RT-PCR).
Results and discussion. The overall prevalence of antibodies (Abs) among all monkeys was low and amounted 11.3% (95% CI: 9.2–13.7%, n = 811) for measles virus, 8.9% (95% CI: 6.2–12.2%, n = 381) for PI type 3 virus, 2.5% (95% CI: 0.8–5.6%, n = 204) for PI type 1 virus, and 7.7% (95% CI: 3.8–13.7%, n = 130) for adenoviruses. When testing 26 autopsy lung samples from monkeys of different species that died from pneumonia, 2 samples from Anubis baboons (Papio аnubis) were positive for of parainfluenza virus type 3 RNA.
Conclusion. Our data suggest the importance of the strict adherence to the terms of quarantine and mandatory testing of monkey sera for the presence of IgM antibodies to the measles virus that indicate the recent infection. The role of PI virus type 3 in the pathology of the respiratory tract in Anubis baboons has been established.
 425-433
425-433


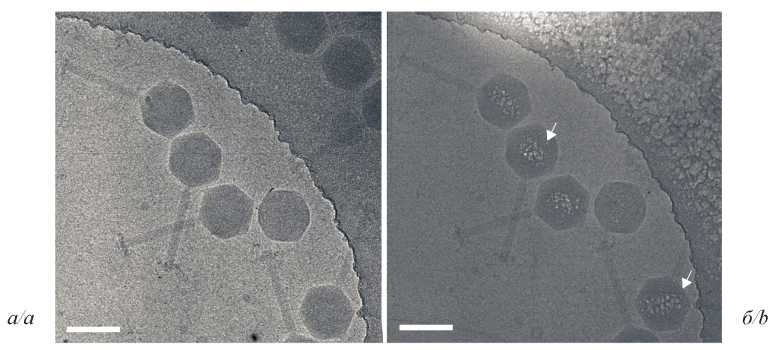
DNA mapping in the capsid of giant bacteriophage phiEL (Caudovirales: Myoviridae: Elvirus) by analytical electron microscopy
Abstract
Introduction. Giant phiKZ-like bacteriophages have a unique protein formation inside the capsid, an inner body (IB) with supercoiled DNA molecule wrapped around it. Standard cryo-electron microscopy (cryo-EM) approaches do not allow to distinguish this structure from the surrounding nucleic acid of the phage. We previously developed an analytical approach to visualize protein-DNA complexes on Escherichia coli bacterial cell slices using the chemical element phosphorus as a marker. In the study presented, we adapted this technique for much smaller objects, namely the capsids of phiKZ-like bacteriophages.
Material and methods. Following electron microscopy techniques were used in the study: analytical (AEM) (electron energy loss spectroscopy, EELS), and cryo-EM (images of samples subjected to low and high dose of electron irradiation were compared).
Results. We studied DNA packaging inside the capsids of giant bacteriophages phiEL from the Myoviridae family that infect Pseudomonas aeruginosa. Phosphorus distribution maps were obtained, showing an asymmetrical arrangement of DNA inside the capsid.
Discussion. We developed and applied an IB imaging technique using a high angle dark-field detector (HAADF) and the STEM-EELS analytical approach. Phosphorus mapping by EELS and cryo-electron microscopy revealed a protein formation as IB within the phage phiEL capsid. The size of IB was estimated using theoretical calculations.
Conclusion. The developed technique can be applied to study the distribution of phosphorus in other DNA- or RNA-containing viruses at relatively low concentrations of the element sought.
 434-441
434-441


Specific features of the pathology of the respiratory system in SARS-CoV-2 (Coronaviridae: Coronavirinae: Betacoronavirus: Sarbecovirus) infected Syrian hamsters (Mesocricetus auratus)
Abstract
Introduction. Verification of histological changes in respiratory system using Syrian (golden) hamsters (Mesocricetus auratus) as experimental model is an important task for preclinical studies of drugs intended for prevention and treatment of the novel coronavirus infection COVID-19.
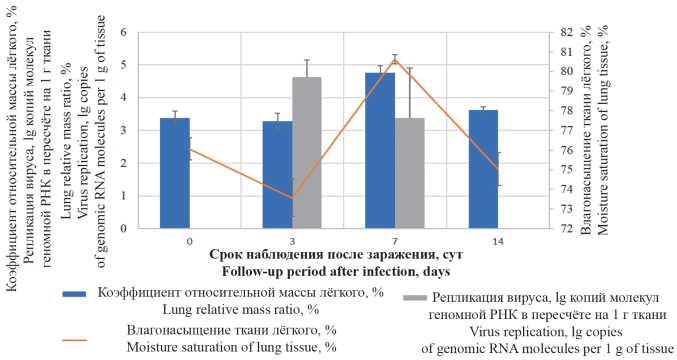
The aim of this work was to study pathological changes of pulmonary tissue in SARS-CoV-2 (Coronaviridae: Coronavirinae: Betacoronavirus; Sarbecovirus) experimental infection in Syrian hamsters. Material and methods. Male Syrian hamsters weighting 80–100 g were infected by intranasal administration of culture SARS-CoV-2 at dose 4 × 104 TCID50/ml (TCID is tissue culture infectious dose). Animals were euthanatized on 3, 7 and 14 days after infection, with gravimetric registration. The viral load in lungs was measured using the polymerase chain reaction (PCR). Right lung and trachea tissues were stained with hematoxylin-eosin and according to Mallory.
Results and discussion. The highest viral replicative activity in lungs was determined 3 days after the infection. After 7 days, on a background of the decrease of the viral load in lungs, a pathologically significant increase of the organ’s gravimetric parameters was observed. Within 3 to 14 days post-infection, the lung histologic pattern had been showing the development of inflammation with a succession of infiltrative-proliferative, edematousmacrophagal and fibroblastic changes. It was found that initial changes in respiratory epithelium can proceed without paranecrotic interstitial inflammation, while in the formation of multiple lung parenchyma lesions, damage to the epithelium of bronchioles and acinar ducts can be secondary. The appearance of epithelioid large-cell metaplastic epithelium, forming pseudoacinar structures, was noted as a pathomorphological feature specific to SARS-CoV-2 infection in Syrian hamsters.
Conclusion. As a result of the study, the specific features of the pathology of the respiratory system in SARSCoV-2 infected Syrian hamsters were described. These findings are of practical importance as reference data that can be used for preclinical studies to assess the effectiveness of vaccines and potential drugs.
 442-451
442-451


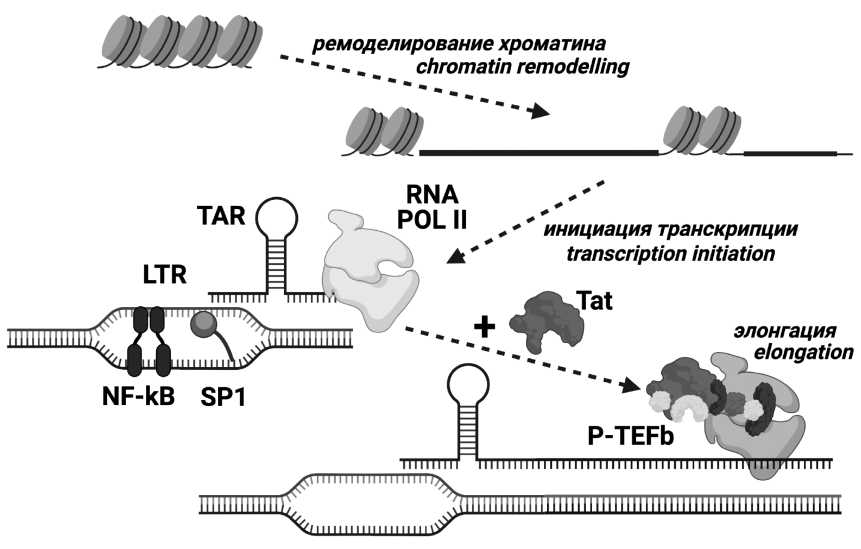
Analysis of Tat protein characteristics in human immunodeficiency virus type 1 sub-subtype A6 (Retroviridae: Orthoretrovirinae: Lentivirus: Human immunodeficiency virus-1)
Abstract
Introduction. Tat protein is a major factor of HIV (human immunodeficiency virus) transcription regulation and has other activities. Tat is characterized by high variability, with some amino acid substitutions, including subtypespecific ones, being able to influence on its functionality. HIV type 1 (HIV-1) sub-subtype A6 is the most widespread in Russia. Previous studies of the polymorphisms in structural regions of the A6 variant have shown numerous characteristic features; however, Tat polymorphism in A6 has not been studied.
Goals and tasks. The main goal of the work was to analyze the characteristics of Tat protein in HIV-1 A6 variant, that is, to identify substitutions characteristic for A6 and A1 variants, as well as to compare the frequency of mutations in functionally significant domains in sub-subtype A6 and subtype B.
Material and methods. The nucleotide sequences of HIV-1 sub-subtypes A6, A1, A2, A3, A4, subtype B and the reference nucleotide sequence were obtained from the Los Alamos international database.
Results and discussion. Q54H and Q60H were identified as characteristic substitutions. Essential differences in natural polymorphisms between sub-subtypes A6 and A1 have been demonstrated. In the CPP-region, there were detected mutations (R53K, Q54H, Q54P, R57G) which were more common in sub-subtype A6 than in subtype B.
Conclusion. Tat protein of sub-subtype A6 have some characteristics that make it possible to reliably distinguish it from other HIV-1 variants. Mutations identified in the CPP region could potentially alter the activity of Tat. The data obtained could form the basis for the drugs and vaccines development.
 452-464
452-464


INFORMATION
 465-468
465-468