Vol 65, No 2 (2020)
- Year: 2020
- Published: 16.05.2020
- Articles: 9
- URL: https://virusjour.crie.ru/jour/issue/view/33
- DOI: https://doi.org/10.36233/0507-4088-2020-65-2
Full Issue
REVIEWS
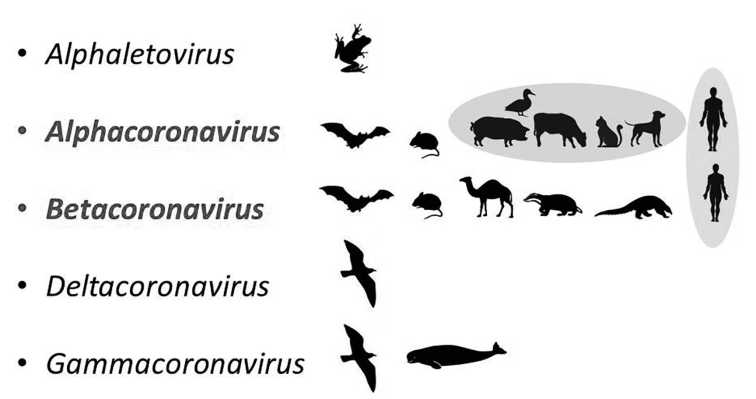
Source of the COVID-19 pandemic: ecology and genetics of coronaviruses (Betacoronavirus: Coronaviridae) SARS-CoV, SARS-CoV-2 (subgenus Sarbecovirus), and MERS-CoV (subgenus Merbecovirus)
Abstract
Since the early 2000s, three novel zooanthroponous coronaviruses (Betacoronavirus) have emerged. The first outbreak of infection (SARS) caused by SARS-CoV virus occurred in the fall of 2002 in China (Guangdong Province). A second outbreak (MERS) associated with the new MERS-CoV virus appeared in Saudi Arabia in autumn 2012. The third epidemic, which turned into a COVID-19 pandemic caused by SARS-CoV-2 virus, emerged in China (Hubei Province) in the autumn 2019. This review focuses on ecological and genetic aspects that lead to the emergence of new human zoanthroponous coronaviruses. The main mechanism of adaptation of zoonotic betacoronaviruses to humans is to changes in the receptor-binding domain of surface protein (S), as a result of which it gains the ability to bind human cellular receptors of epithelial cells in respiratory and gastrointestinal tract. This process is caused by the high genetic diversity and variability combined with frequent recombination, during virus circulation in their natural reservoir - bats (Microchiroptera, Chiroptera). Appearance of SARS-CoV, SARS-CoV-2 (subgenus Sarbecovirus), and MERS (subgenus Merbecovirus) viruses is a result of evolutionary events occurring in bat populations with further transfer of viruses to the human directly or through the intermediate vertebrate hosts, ecologically connected with bats.
This review is based on the report at the meeting «Coronavirus - a global challenge to science» of the Scientific Council «Life Science» of the Russian Academy of Science: Lvov D.K., Alkhovsky S.V., Burtseva E.I. COVID-19 pandemic sources: origin, biology and genetics of coronaviruses of SARS-CoV, SARS-CoV-2, MERS-CoV (Conference hall of Presidium of RAS, 14 Leninsky Prospect, Moscow, Russia. April 16, 2020)
 62-70
62-70


Prion diseases and the biosecurity problems
Abstract
 71-76
71-76


ORIGINAL RESEARCH
Ancient variants of the Epstein–Barr virus (Herpesviridae, Lymphocryptovirus, HHV-4): hypotheses and facts
Abstract
Introduction. Molecular studies have shown that viruses appeared in the early stages of the evolution of life. For millions of years, viruses have evolved by changing old and acquiring new sequences in their RNA or DNA. It is assumed that most viruses have common ancestors. Such an ancestor, an ancient strain, probably existed for Epstein-Barr virus (EBV) as well.
Aim. To find out whether ancient strains of EBV persist in modern Russian ethnic groups today.
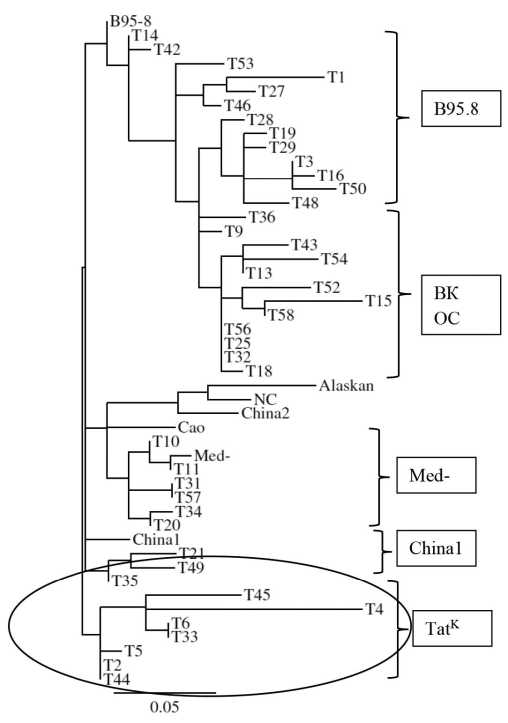
Material and methods. The object of the study was the EBV LMP1oncogene, which is most suitable for molecular genetic analysis. LMP1 was amplified from the oral cavity washings obtained from representatives of two ancient ethnic groups of Russia - Tatars and Slavs. The LMP1 amplicons were sequenced in both directions; their nucleotide sequences translated into amino acid (LMP1) were evaluated using the classification suggested by Edwards et al. 1999. To establish genetic relationships between LMP1 variants, a phylogenetic tree was constructed by the neighbor-joining method using the MEGA software package.
Results and discussion. Analysis of LMP1 sequences from washings of the Slavs oral cavity demonstrated the presence of LMP1 variants with varying degrees of transforming potential: B98.5/A, China1, Med-, and NC. The analysis of LMP1 sequences from washings of Tatar oral cavity also made it possible to identify oncoprotein variants such as B95.8/A, China1, Med-, as well as a group of variants out of classifications (LMP1-OK). An important finding was the identification of 7 variants of LMP1 from Tatars, designated as LMP1-TatK, that contained two unique deletions of 5 aa in codons 312-316 and 382-386, which were absent in the LMP1 variants from Slavs and from previously examined cancer patients and healthy individuals, as well as in sequences from open computer databases. The uniqueness of the LMP1-TatK variant is confirmed both by phylogenetic analysis of LMP1 sequences of Tatar origin and by the analysis of 11 aa repeats and 5 aa insertions in the C-terminal region of the oncoprotein. The morbidity and mortality rates from neoplasms, including EBV-associated pathologies, did not differ significantly between two studied ethnic groups infected with different EBV strains.
Conclusion. The data obtained allowed us to suggest that: 1) LMP1-TatK could be refered to an evolutionarily ancient EBV strain that persists among Tatars and; 2) LMP1-TatK belongs to the so-called “Volga” EBV virus strain, the common strain among the population of the Volga region. Extended studies of EBV isolates from residents of this region may probably shed the light on the origin of LMP1-TatK.
 77-86
77-86


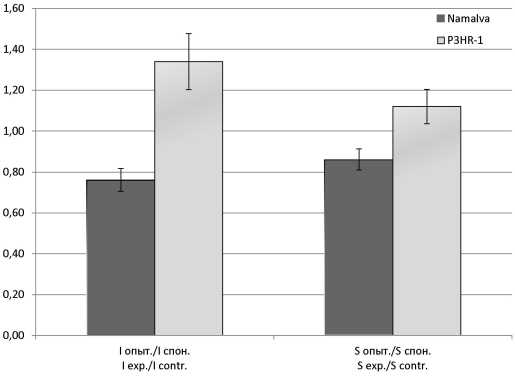
Interferon-regulating activity of the CelAgrip drug and its influence on the formation of reactive oxygen species and expression of innate immunity genes in Burkitt’s lymphome cell cultures
Abstract
Introduction. Interferons (IFN) and IFN inducers are effective in suppressing viral reproduction and correcting of the innate immunity mechanisms.
The aim of the study was to test the hypothesis of the possible involvement of the IFN inducer CelAgrip (CA) as an activator or suppressor of antiviral effects in Burkitt’s lymphoma (LB) cell cultures with different ability to produce Epstein-Barr virus antigens (EBV).
Material and methods. The kinetic analysis of the dynamics of reactive oxygen species (ROS) production and determination of gene group expression by real-time PCR in response to CA treatment were done in human cell lines LB P3HR-1 and Namalva, spontaneously producing and not producing EBV antigens.
Results and discussion. When treating CA in Namalva cells, a decrease in the ROS activation index was found; in P3HR-1 cells, an increase was observed. After treatment with CA, there was no reliable activation of the IFN-α, IFN-β and IFN-λ genes in Namalva cells, but the expression of the ISG15 and P53(TP53) genes was increased more than 1200 times and 4.5 times, respectively. When processing the CA of P3HR-1 cells, the expression of IFN-α genes increased by more than 200 times, IFN-λ - 100 times, and the ISG15 gene - 2.2 times. The relationship between IFN-inducing action of CA and the activity of ISG15 and ROS in LB cell cultures producing and not producing EBV antigens is supposed.
Conclusion. In Namalva cells that do not produce EBV antigens the treatment of CA results in suppression of ROS generation and activation of the expression of genes ISG15 and P53 (TP53); in P3HR-1 cells producing EBV antigens, the opposite picture is observed - the formation of ROS and the expression of the IFN-α and IFN-λ genes are activated and the activity of the ISG15 and P53 (TP53) genes is suppressed.
 87-94
87-94


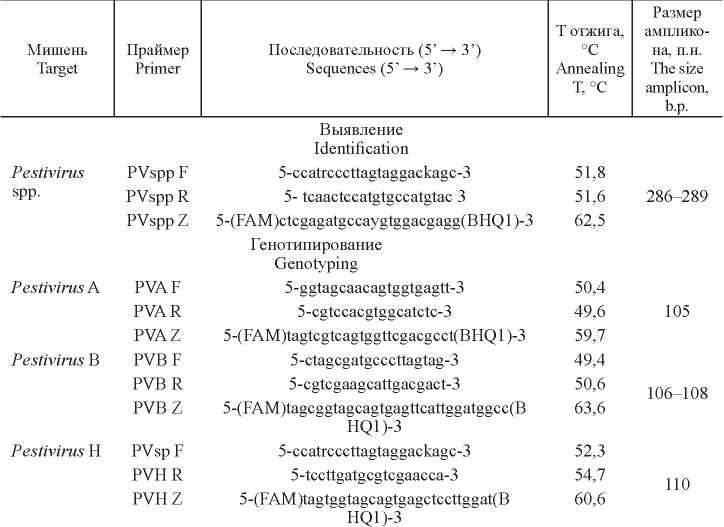
Detection of bovine pestiviruses by a multiplex real-time polymerase chain reaction
Abstract
Introduction. Pestiviruses are the cause of reproductive problems, diseases of the gastrointestinal and respiratory tracts of animals. Three species are important for cattle: Pestivirus A, B, and H. Fast and reliable methods of differentiation of these pathogens are currently needed.
Aims and objectives of the study: the development of multiplex real time PCR for the simultaneous detection and differentiation of three viruses.
Material and methods. The nucleotide sequences of the conserved regions of the 5´-UTR genes of pestiviruses A, B, and H served as a target.
Results. The reaction showed high specificity, sensitivity, reproducibility and was able to detect virus RNA at a concentration of not less than 0.6-1.2 lg TCID50/cm3. Cross-reactions with other pestiviruses were not observed. Real time PCR confirmed the results obtained previously in RT-PCR with gel electrophoresis detection. In a parallel study of 1823 biological samples, the results of the two reactions were completely consistent. Pestivirus spp. was detected in 76 samples, Pestivirus A was present in 73 samples, Pestivirus B - in 3 samples, and Pestivirus H was not detected.
Discussion. A two-step real time PCR was developed for the simultaneous detection and differentiation of three pestiviruses. Modified pan primers of S. Vilcek et al. were used for the first reaction, and primers and probes of our own design were used for virus typing, which resulted in high reaction efficiency.
Conclusion. On the big dairy farms for livestock maintenance, there are favorable conditions for the circulation of pathogenic viruses. In this situation, rapid diagnostic methods are needed to quickly identify of several viruses. Real-time triplex analysis can be recommended as the rapid method for mass epidemiological studies, as well as for screening fetal calf serum used for virus cultivation in medicine and veterinary practice.
 95-102
95-102


In silico prediction of B- and T-cell epitopes in the CD2v protein of african swine fever virus (African swine fever virus, Asfivirus, Asfarviridae)
Abstract
Introduction. African swine fever virus (ASF) is a large DNA virus that is the only member of the Asfarviridae family. The spread of the ASF virus in the territory of the Russian Federation, Eastern Europe and China indicates the ineffectiveness of existing methods of combating the disease and reinforces the urgent need to create effective vaccines. One of the most significant antigens required for the formation of immune protection against ASF is a serotype-specific CD2v protein.
The purpose of the study. This study presents the results of immuno-informatics on the identification of B- and T-cell epitopes for the CD2v protein of the ASF virus using in silico prediction methods.
Material and methods. The primary sequence of the CD2v protein of the ASFV virus strain Georgia 2007/1 (ID-FR682468) was analyzed in silico by programs BCPred, NetCTLpan, VaxiJen, PVS and Epitope Conservancy Analysis.
Results. Using the BCPred and VaxiJen programs, 4 major B-cell immunogenic epitopes were identified. Analysis of the secretory region of ASF virus CD2v protein in NetCTLpan revealed 5 T-cell epitopes from the 32nd to the 197th position of amino acids that cross-link from the 1st to the 13th allele of the MHC-I of pig
Discussion. This study presents the results in silico prediction to identify B- and T-cell epitopes of ASF virus CD2v protein. The soluble region of the CD2v protein can be included in the recombinant polyepitope vaccine against African swine fever.
Conclusion. B- and T-cell epitopes in the secretory region of the CD2v protein (from 17 to 204 aa) of ASF virus were identified by in silico prediction. An analysis of the conservatism of the identified B- and T-cell epitopes allowed us to develop a map of the distribution of immune epitopes in the CD2v protein sequence.
 103-112
103-112


TO VIROLOGIST’S AID
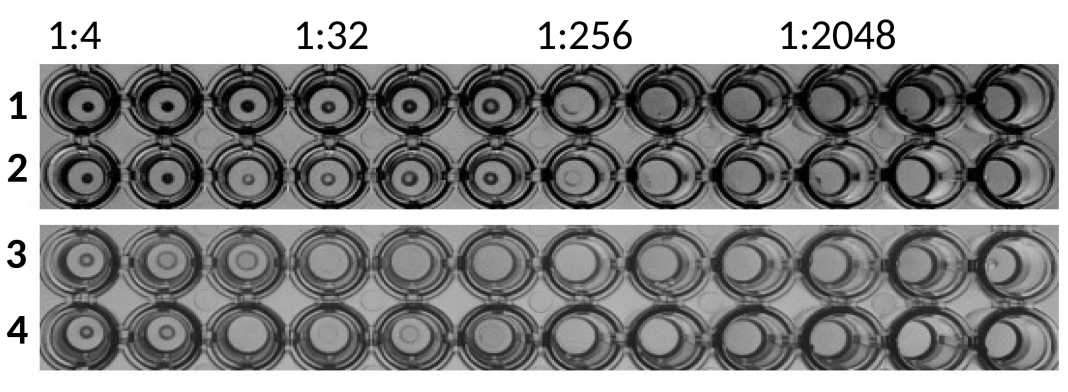
Method for evaluating the neuraminidase activity of receptor-destroying enzyme (RDE) compounds using the influenza virus
Abstract
Introduction. The classic hemagglutination inhibition reaction (RTGA) is used to determine the level of antiviral antibodies in human and animal serum specimens. During the performance of RTGA the tested sera must be treated with a receptor-destroying enzyme (RDE) to remove serum glycans that degrade the accuracy of the RTGA results. To optimize the amounts of RDE compounds used, it is necessary to know their real neuraminidase activity. This article describes a simple and economical method for testing the neuraminidase activity of receptor-destroying compounds using standard reagents and laboratory equipment.
Aims of investigation. Design of an improved simple and convenient method for evaluating the neuraminidase activity using the flu virus.
Material and methods. Here, we propose a convenient method for evaluating the activity of neuraminidase by double-fold dilution procedure with human or animal erythrocytes followed by hemagglutination assay with influenza A virus.
Results and discussion. The method is based on the ability of neuraminidase to hydrolyze sialic acid residues on the cell surface of erythrocytes, that deprives red blood cells to be agglutinated with the flu virus, since these sialic glycans provide virus attachment and hemagglutination.
Conclusion. The designed method allows the accurate measurement of the receptor-destroying (neuraminidase) activity of RDE compounds and the comparison of the compounds with each other. This test is necessary to optimize the RTGA protocol when monitoring blood sera of animals and humans after influenza infection and/or Acute Respiratory diseases (ARD). The designed method can be included in the guidelines of regulations for the RTGA protocol, which is used in different laboratories to monitor the epidemic process of influenza and ARD infections.
 113-118
113-118


BOOK REVIEW
F.I. Ershov «History of virology from D.I. Ivanovsky to the present day»
Abstract
 119-119
119-119


OBITUARY
 120-120
120-120