Avian flu: «for whom the bell tolls»?
- Authors: Zhirnov O.P.1,2, Lvov D.K.1
-
Affiliations:
- The D.I. Ivaovsky Institute of Virology, The N.F. Gamaleya Research Center of Epidemiology and Microbiology, The Russian Ministry of Health
- The Russian-German Academy of Medical-Social and Biotechnological Sciences, Skolkovo Innovation Center
- Issue: Vol 69, No 2 (2024)
- Pages: 101-118
- Section: EDITORIAL CONCEPT
- URL: https://virusjour.crie.ru/jour/article/view/16636
- DOI: https://doi.org/10.36233/10.36233/0507-4088-213
- EDN: https://elibrary.ru/gtekdx
- ID: 16636
Cite item
Abstract
The family Orthomyxoviridae consists of 9 genera, including Alphainfluenza virus, which contains avian influenza viruses. In two subtypes H5 and H7 besides common low-virulent strains, a specific type of highly virulent avian virus have been described to cause more than 60% mortality among domestic birds. These variants of influenza virus are usually referred to as «avian influenza virus». The difference between high (HPAI) and low (LPAI) virulent influenza viruses is due to the structure of the arginine-containing proteolytic activation site in the hemagglutinin (HA) protein. The highly virulent avian influenza virus H5 was identified more than 100 years ago and during this time they cause outbreaks among wild and domestic birds on all continents and only a few local episodes of the disease in humans have been identified in XXI century. Currently, a sharp increase in the incidence of highly virulent virus of the H5N1 subtype (clade h2.3.4.4b) has been registered in birds on all continents, accompanied by the transmission of the virus to various species of mammals. The recorded global mortality rate among wild, domestic and agricultural birds from H5 subtype is approaching to the level of 1 billion cases. A dangerous epidemic factor is becoming more frequent outbreaks of avian influenza with high mortality among mammals, in particular seals and marine lions in North and South America, minks and fur-bearing animals in Spain and Finland, domestic and street cats in Poland. H5N1 avian influenza clade h2.3.4.4b strains isolated from mammals have genetic signatures of partial adaptation to the human body in the PB2, NP, HA, NA genes, which play a major role in regulating the aerosol transmission and the host range of the virus. The current situation poses a real threat of pre-adaptation of the virus in mammals as intermediate hosts, followed by the transition of the pre-adapted virus into the human population with catastrophic consequences.
Full Text
Avian flu: «For whom the bell tolls»?*
The increase in avian influenza-related adverse and threatening events worldwide determines the need to revisit the debate on this issue.
General characterization of the Orthomyxoviridae family
The Orthomyxoviridae family according to the modern classification is divided into 9 genera: Alpha-, Beta-, Delta-, Gamma-, Thogoto-, Quaranja-, Mycis-, Sardino- and Isainfluenza viruses (Table 1) [1]. The family is formed by viruses that have an outer lipoprotein envelope (enveloped viruses), single-stranded RNA of negative polarity in the form of 7 or 8 segments (in different genera) encoding up to 16 unique proteins [2]. Orthomyxoviruses have a wide range on the planet in all three environments: aquatic, air and land and have tropism to a wide range of organisms – arthropods, fish, birds and mammals, including humans, as seen in Table 1 [3]. In populations of birds, the main natural reservoir of influenza A virus, infection occurs via the fecal-oral route.
Table 1. Classification of the family Orthomyxoviridae
Таблица 1. Классификация семейства Orthomyxoviridae
Virus genus number Номер рода вирусов | Genus name Наименование рода | Host spectrum of the virus Спектр хозяев вируса |
1. 2. 3. 4. | Genus: Alphainfluenzavirus (Flu A) Genus: Betainfluenzavirus (Flu B) Genus: Deltainfluenzavirus (Flu C) Genus: Gammainfluenzavirus (Flu D) | Birds (Aves)/Птицы, Human (Homo sapiens)/Человек, Mammalians (Mammalia)/Млекопитающие |
5. 6. | Genus: Thogotovirus Genus: Quaranjavirus | Arthropods (Arthropoda)/Членистоногие: Ticks (Ixodidae)/Клещи иксодовые, Argasid mites (Argasidae)/Клещи аргасовые |
7. 8. 9. | Genus Mykissvirus Genus: Sardinovirus Genus: Isavirus | Fishes (Ichthya)/Рыбы |
Note. Classification of the family Orthomyxoviridae in accordance with the recommendations of ICTV-2022 [1].
Примечание. Классификация семейства Orthomyxoviridae в соответствии с рекомендациями ICTV-2022 [1].
The peculiarity of orthomyxoviruses is their high variability, which is due to the functional properties of viral RNA polymerase and the segmented structure of the viral genome. Viral RNA polymerase provides synthesis and replication of the viral genome, which it carries out with a large number of random errors, amounting to about one mutation per 104 nucleotides of the genome per one read. For comparison, we note that human DNA-dependent RNA polymerase II allows an error of one mutation only during polymerization of 108 nucleotides in the synthesized mRNA molecule [4]. The high error rate of viral polymerase generates high variability of the virus and the formation of a large number of viral variants, also known as viral quasispecies, in the viral progeny. The genetic diversity of the viral population allows the virus to cross the inter-taxon barrier to a new host. This mechanism of gradual accumulation of mutations of adaptive character has been called viral drift. Importantly, drift is based on a dual mechanism that regulates (1) adaptation of the virus to host organism factors and (2) overcoming its immune response. As a result of interaction at the molecular-genetic level between the virus and host populations, the formation of the virus population gene pool occurs [5]. The period of its formation takes place over millions of years, closely interacting with the elements of the biosphere under changing environmental conditions.
The second mechanism ensuring high natural variability of orthomyxoviruses is the exchange (reassortment) of genomic RNA segments between viruses adapted to different animal species [6, 7]. The crossing of such adaptive viral variants underlies the phenomenon of genetic shift, leading to the emergence of radically new viral reassortants and unique antigenic and functional variants of influenza virus, which have the potential for trans-taxon transfer transfer of the virus from one species of macroorganism to another and changing the range of virus hosts and the formation of new pandemic variants of the virus.
Two levels of influenza A virulence (Alphainfluenzavirus genus)
The high variability of influenza A virus determines the formation of a massive population gene pool and the distribution in nature of a wide range of viral variants with a pronounced diversity of pathogenic and antigenic properties [8–10]. The Alphainfluenzavirus genus (influenza A viruses) consists of 18 antigenic subtypes of hemagglutinin (HA) numbered H1–H18 and 11 subtypes of neuraminidase (NA) protein numbered N1–N11. According to this numbering, the antigenic subtypes HA and NA are indicated in the name of the variant (strain) of influenza viruses [11–13].
Among the 18 known subtypes of the Alphainfluenzavirus genus, viruses of subtypes H5 and H7 are of special interest. The populations of these viral subtypes consist of two biological species of viruses with low (LPAI) and high (HPAI) virulence for host macroorganisms [13, 14]. Low pathogenic avian influenza causes a localized infectious process, usually of the respiratory or intestinal tract of the host often without clinically evident symptoms, with a population mortality rate of less than 1% [15]. Highly pathogenic viruses of subtypes H5 and H7, which received their original historical name in the late 19th century as «fowl plaque» or «avian typhus», cause massive viremia and dissemination of the virus in the host organism in most susceptible, usually domestic and farm animals, with the development of generalized (pantropic) forms of infection with high lethality close to 100% [16]. In avian populations, the main natural reservoir of influenza A virus, infection occurs by fecal-oral route. The biological system «influenza A virus – birds» has probably functioned in the biosphere for more than 100 million years since the Cretaceous period of the Mesozoic era [5]. Birds are a natural reservoir, the arena of recombinant processes as a result of interpopulation contacts arising during migration periods, especially in migration hubs where bird populations from different parts of the range accumulate in large numbers [17–20]. The main natural reservoirs of influenza A viruses are species of the goose family (Anseriformes), duck family (Anatidae), rye family (Charadriiformes), waders (Scolopatidae), gulls (Laridae) and others. In nature, influenza A viruses have been isolated from more than 100 species of birds (12 orders, 25 families, more than 50 genera) [5, 21–23]. The first highly pathogenic influenza virus of subtype H5 was isolated in Scotland in 1959 from chickens – strain A/chicken/Scotland/1959 (H5N1) [24], and then in South Africa during an epizootic among migrating Arctic terns Sterna macrura – strain A/tern/South Africa/61 (H5N3) [25]. In the 1970s, the concept of natural focality of influenza A virus was formed (Fig. 1) [5, 8, 18, 21, 22, 26–29]. It should be taken into account that birds are the main carrier and reservoir in nature of highly pathogenic variety of representatives of subtype H5N1 of the Alphainfluenzavirus genus [28].
Fig. 1. Founders of the concept of natural focality of the Influenza A virus (Orthomyxoviridae: Influenza A virus). Pictured from left to right: Graham Laver (Australia), Dmitry Lvov (USSR), Robert Webster (USA) [8, 22, 26, 27].
Рис. 1. Основоположники концепции природной очаговости вируса гриппа А (Orthomyxoviridae: Influenza A virus). На фото слева направо: Грэм Лавер (Австралия), Дмитрий Львов (СССР), Роберт Вебстер (США) [8, 22, 26, 27].
The molecular basis of the cardinal difference in virulence properties of influenza A virus is determined by the structure of the site (the so-called proteolytic site) of viral hemagglutinin HA0 (mol.wt. 75 kDa), where proteolytic cleavage into two subunits HA1 (55 kDa) and HA2 (20 kDa) occurs, which retain a disulfide bond after cleavage (Fig. 2). This cleavage of the viral hemagglutinin protein activates its lipid membrane fusion function (symplast formation function), making the virion infectious and capable of infecting new target cells [30–33]. The phenomenon of influenza virus virulence enhancement through proteolysis of HA0 into HA1+HA2 is called proteolytic activation of the virus [33, 34].
Fig. 2. Structure of the proteolytic activation site of the influenza A virus hemagglutinin (HA) protein.
The HA protein site in the junction zone of HA1 (55 kDa) and HA2 (20 kDa) subunits formed after cleavage of the precursor molecule, HA0 (75 kDa), is shown. Low pathogenic viruses of subtypes H1–H18 contain a single arginine (Arg) residue (panel A), whereas highly pathogenic subtypes H5 and H7 have several arginine and lysine (Lys) residues present in the proteolysis site, the so-called polyarginine site of proteolysis (panel B). The polyarginine site is highly sensitive to a wide range of cellular proteases and is readily cleaved in the vast majority of target cell types [16, 33, 35–37].
Рис. 2. Структура сайта протеолитической активации белка гемагглютинина (НА) вирусов гриппа А.
Показан участок белка НА в зоне соединения субъединиц НА1 (55 кДа) и НА2 (20 кДа), формирующихся после расщепления молекулы-предшественника НА0 (75 кДа). Слабопатогенные вирусы субтипов Н1–Н18 содержат единичный остаток аргинина (Arg) (панель А), тогда как у высокопатогенных субтипов Н5 и Н7 в участке протеолиза присутствуют несколько остатков аргинина и лизина (Lys), так называемый полиаргининовый сайт протеолиза (панель Б). Полиаргининовый сайт имеет высокую чувствительность к широкому спектру клеточных протеаз и легко расщепляется в подавляющем большинстве типов клеток-мишеней [16, 33, 35–37].
In low pathogenic viruses, the HA proteolytic site consists of a single arginine residue (Arg), the so-called «monoarginine site», whereas in highly pathogenic influenza viruses it consists of several arginine residues and possibly lysine (Lys), the so-called «polyarginine site». The cleavage of monoarginine HA0 is carried out by trypsin-like proteases and, as a rule, occurs post-translationally already on the cell plasma membrane when the virion leaves the infected cell. The efficiency of such cleavage in infected cells was about 30–40% of the molecules of synthesized HA0 protein [35, 36]. In highly pathogenic viruses of subtypes H5 and H7, cleavage of HA0→HA1+HA2 occurs inside infected cells contranslationally during the process of the HA0 molecule synthesis and the virus leaves the cell in the infectious form with cleaved HA1+HA2 protein. HA0 cleavage of such viruses is performed by intracellular ubiquitin proteases of furin series on the membranes of the Golgi apparatus with high efficiency, close to 100% of HA0 molecules [37]. As a result of intensive proteolysis HA0→HA1+HA2, the synthesized highly pathogenic viruses easily pass through intercellular contacts and basal membranes in the primary infectious focus of the respiratory and/or intestinal tracts, penetrate into the bloodstream and cause generalized pantropic infection of the macroorganism [16, 33]. Low pathogenic viruses due to partial cleavage of monoarginine site in HA0 and not so effective activation of infectivity and fusion properties of virions have low ability to overcome protective cellular barriers in the host organism. As a rule, such viral strains, which include LPAI viruses and seasonal human influenza viruses, induce a local pathological process in the primary organ and do not cause multi-organ dissemination of the virus and a generalized form of the infectious process in the affected organism [33, 37–39].
Factors regulating the host range of the virus
An important feature of influenza A virus is its ability to infect a wide range of animals, including birds (the main natural reservoir of influenza A virus) and humans (Fig. 3).
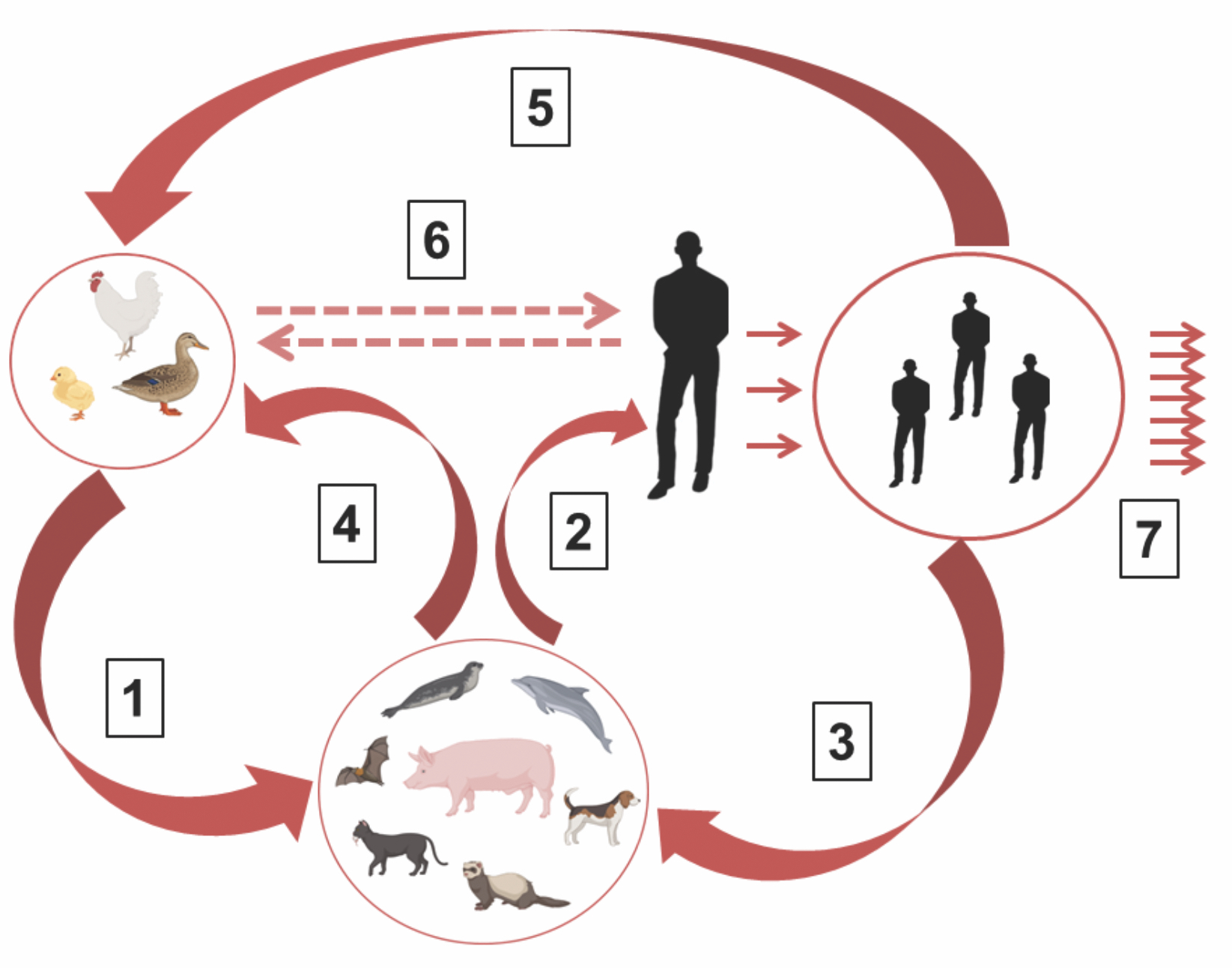
Fig. 3. Adaptation stages of the H5N1 avian influenza virus and the emergence of a pandemic variant of the human influenza plague virus.
The H5N1 avian influenza virus can be transmitted directly to humans (direct transmission route) or indirectly, first to mammals such as pigs, fur-bearing animals, animals of prey, seals, etc., followed by an indirect transmission route to humans). In the human body (with direct transmission) and mammals, the avian virus undergoes partial adaptation to the human body through adaptive mutations or reassortation of viral genomic RNA segments between avian and human influenza viruses. The H5N1 avian influenza virus, partially adapted to humans, can return to the population of birds and mammals and form a natural reservoir of a permanent potential threat of the emergence of highly pathogenic epidemic virus (HPAV) subtypes H5 and H7 and its subsequent return to the human population. Designations: 1 ‒ Initial adaptation of influenza virus to humans (mutations in the NP, PB2, HA, NA genes and others); 2 ‒ Partial adaptation of virus to human; 3 ‒ Maintenance of interspecies virus transmissions in biosphere; 4 ‒ Virus transmission from mammals to birds; 5 ‒ Maintenance of interspecies transmissions in biosphere; 6 ‒ Direct transmission way from birds to humans and back; 7 ‒ Development of epidemic process.
Рис. 3. Этапы адаптации вируса птичьего гриппа H5N1 и возникновения пандемического варианта вируса.
Птичий вирус гриппа H5N1 способен непосредственно переходить на человека (прямой путь передачи) или опосредованно ‒ сначала на млекопитающих животных, таких как свиньи, пушные звери, хищные звери, тюлени и др., с последующим переходом на людей (непрямой путь передачи). В организме человека и млекопитающих животных птичий вирус претерпевает частичную адаптацию к организму человека посредством адаптационных мутаций либо реассортацией сегментов вирусной геномной РНК между вирусами гриппа птиц и человека. Частично адаптированный к человеку вирус птичьего гриппа H5N1 может возвращаться в популяцию птиц и животных и формировать природный резервуар перманентной потенциальной угрозы возникновения эпидемического вируса высокопатогенного вируса человека субтипов H5 и H7. Обозначения: 1 ‒ первичная адаптация вируса гриппа к человеку (мутации в генах NP, PB2, HA, NA и др.); 2 ‒ частичная адаптация вируса к человеку; 3 ‒ поддержание межвидовой передачи вируса в биосфере; 4 ‒ передача вируса от млекопитающих птицам; 5 ‒ поддержание межвидовой передачи в биосфере; 6 ‒ прямой путь передачи инфекции от птиц к человеку и обратно; 7 ‒ развитие эпидемического процесса.
As an obligate parasite of the host cell, the influenza virus interacts with cellular factors and has structural patterns in viral proteins for this purpose, also known as genetic signatures. For successful reproduction, these structural domains (patterns) must be concordant with the cells of a particular host species. When the host changes, the virus needs to change these patterns and adapt to the new host species.
In the replication process of influenza A virus, 7 main points (stages) of interaction with the host cell apparatus can be distinguished:
The above stages involve various viral proteins that regulate the interaction with the cell giving rise to new viral progeny and determine the development of the infectious process in the affected organism [38].
Table 2. Comparison of amino acid patterns of NP protein in influenza A viruses isolated from mammals and birds
Таблица 2. Сравнение аминокислотных паттернов белка NP у вирусов гриппа А, изолированных от млекопитающих и птиц
Subtype and origin of the virus (type of signature)1 Субтип и происхождение вируса (вид штампа)1 | Numbers of amino acid positions in the genetic signature of the NP protein2 Номера позиций аминокислот в генетическом штампе белка NP2 | Matching positions in the human virus NP signature, % Совпадения позиций в штампе NP вируса человека, % | Matching positions in the avian virus NP signature, % Совпадения позиций в штампе NP вируса птиц, % | ||||||||||||||
16 | 33 | 61 | 100 | 109 | 214 | 283 | 293 | 305 | 313 | 357 | 372 | 422 | 442 | 455 | |||
H1N1 human (classic) H1N1 человека (классический) | D | I | L | V | V | K | P | K | K | Y | K | D | K | A | E | 100 | 0 |
H1N1 minks H5N1 норки | G/D3 | I | I | R | V | R | L | R | R | F | Q | E | R | T | D | 13 | 87 |
H5N1 avian H5N1 птичий | G | V | I | R | I | R | L | R | R | F | Q | E | R | T | D | 0 | 100 |
Note. 1 ‒ primary NP protein sequences were taken from GenBank and GISAID databases with 10–15 strains for each group; 2 ‒ the numbering of amino acid positions begins with the N-terminal methionine in the NP protein molecule (70 kDa; the total sequence of 498 amino acids); 3 ‒ the marked position has heterogeneity in viral populations, in which the amino acid D is identified in 10‒20% of viral isolates.
Примечание. 1 ‒ первичные последовательности белка NP взяты из баз данных GenBank и GISAID по 10‒15 штаммов для каждой группы; 2 ‒ нумерация позиций аминокислот начинается с N-концевого метионина в молекуле белка NP (70 кДа; общая протяженность 498 аминокислот); 3 ‒ отмеченная позиция имеет гетерогенность в вирусных популяциях, в которых аминокислота D обнаруживается в 10‒20% вирусных изолятов.
The proteins NP, PB2, HA, and NA make the most significant contribution to the regulation of virus tropism and infectivity to the host (Table 2). The main protein of the viral nucleocapsid NP plays a major role in overcoming the host innate immunity due to the Mx factor [40]. The NP protein of avian influenza viruses is recognized by human nuclear Mx factor, which binds the viral nucleocapsid into polymeric complexes and stops virus reproduction [41]. In contrast, human NP viruses are not recognized by human Mx factor; the virus eludes the human immune response and successfully maintains its replication in the human body. In addition to Mx factor recognition, the role of NP protein in host circle regulation is due to its interaction with the cellular nuclear factors ANP32A and ANP32B, which are involved in the intranuclear process of histone acetylation and cytoplasmic transport of captioned viral mRNA, as well as in blocking the antiviral mitochondrial protein MAVS [42–45], alpha-importin regulating the transport of viral RNP through the nuclear pore in the infected cell [46, 47], cellular factor UAP56 involved in splicing of nuclear RNAs [48]. Genetic patterns of human NP virus protein include key amino acids at positions: 16/D; 33/I; 61/L; 100/I/V; 109/V; 214/K; 283/P; 293/K; 305/K; 313/Y; 357/K; 357/K; 372/D; 422/K; 442/A; 455/E; whereas in avian influenza viruses, this amino acid pattern has distinct differences (Table 2) [49–54]. Consequently, in the case of avian influenza viruses transferring from birds to humans, the amino acid pattern must change and have a human algorithm to overcome the Mx barrier and successfully replicate the viral genome in the cell nucleus.
Table 3. Mutations that control the host range of the H5N1 influenza virus
Таблица 3. Мутации, контролирующие круг хозяев вируса гриппа H5N1
Viral gene/protein Вирусный ген/белок | Viral protein functions that determine association with the host Функции вирусного белка, определяющие связь с хозяином | Mutations leading to adaptation in mammals Мутации, ведущие к адаптации у млекопитающих |
NP | Overcomes the MxA factor of human innate immunity Regulates the intranuclear transport of viral RNP through the pores of the cell nucleus Interacts with nuclear RNA helicase UAP56, which is involved in the splicing and transport of intranuclear pro-mRNAs, and also blocks the antiviral mitochondrial protein MAVS, interacts with cellular caspases, factors TRIM4, TRIF3, Rab11a, eEF1D, etc. Преодолевает фактор МхА врожденного иммунитета человека Регулирует внутриядерный транспорт вирусного РНП через поры клеточного ядра Взаимодействует с ядерной РНК хеликазой UAP56, участвующей в сплайсинге и транспорте внутриядерных про-мРНК, а также блокирует антивирусный митохондриальный белок MAVS, взаимодействует с клеточными каспазами, факторами TRIM4, TRIF3, Rab11a, eEF1D и др. | 16/D; 33/I; 61/L; 100/I/V; 109/V; 214/K; 283/P; 293/K; 305/K; 313/Y; 357/K; 357/K; 372/D; 422/K; 442/A; 455/E |
PB2 | Recognizes and hydrolyzes “capped” cellular mRNAs during transcription of the viral genome in the cell nucleus Interacts with human nuclear factors ANP32A and ANP32B Распознает и гидролизует «кэпированные» клеточные мРНК при транскрипции вирусного генома в ядре клеток Взаимодействие с ядерными факторами ANP32A и ANP32B человека | 271/A; 526/K 590/S; 627/K; 701/N |
HA | Determines adsorption on SA receptors of human cell types 2‒6 Forms a functional complex with the viral protein NA for the specific hydrolysis of SA residues of type 2‒6 bonds on the surface receptors of host cells Regulates the contagiousness of the virus Определяет адсорбцию на SA рецепторах 2‒6-го типа клеток человека Формирует функциональный комплекс с вирусным белком NA специфического гидролиза остатков SA 2‒6-го типа связи на поверхностных рецепторах клеток хозяина Регулирует контагиозность вируса | 137/A; 155/T; 160/A; 190/D; 193/R; 225/D; 226/L; 228/S |
NA | Recognizes and hydrolyzes SA residues with the α2-6 bond type on cellular receptors and in respiratory mucin Participates in the regulation of virus contagiousness Распознает и гидролизует остатки SA с типом связи α2-6 на клеточных рецепторах и в муцине респираторного секрета Участвует в регуляции контагиозности вируса | Mutations/Мутации: 46/D; 74/S; 151/D; 163/L; 204/M; 369I; 396M; 420/G No deletion in the stalk and the lack of a 2nd receptor center in the NA/ Отсутствие делеции в ножке и 2-го рецепторного центра в NA |
Note. Description of mutations and corresponding links to publications are given in the text of the article.
Примечание. Описание мутаций и соответствующие ссылки на публикации приведены в тексте статьи.
The polymerase protein PB2 functions in cooperation with the NP protein (Table 3). In this protein PB2, the positions of amino acids T271A, E627K, and D701N are important for adaptation to the human body and efficient synthesis of viral RNAs, they differ in avian influenza viruses [42]. Amino acid substitutions at these positions in avian viruses increase affinity to human cellular factors, leading to enhanced replication of such viral variants during human infection [54, 55]. When performing their virus-specific functions of RNA synthesis, NP and PB2 proteins interact with host factors at the stages of intranuclear replication of the virus genome (see stages 3–5 listed above).
The second most important group of host adaptation factors is the tandem of the influenza virus external receptor proteins HA and NA. These proteins determine the stability and spread of the virus in the external environment and control the specificity of receptor recognition on the epithelial surface at the initial stage of infection during host infection, as well as cellular glycoproteins in the budding zone at the final stage of assembly of newly synthesized virions on the surface of infected cells. These proteins function in cooperation [56, 57] and there is a reciprocal balance of their functions, which plays an important role in the adaptation of the virus to the host [58, 59].
In avian influenza viruses, the receptor and enzymatic glycosidase functions of HA and NA proteins are realized preferentially through interaction with glycoprotein receptors with α2-3 terminal sialic acid (SA) residues linked to galactose residues, which are enriched in the respiratory and intestinal tracts of most birds. In human viruses, the HA–NA protein tandem has a preferential affinity for glycoproteins with a terminal SA residue linked to galactose by an α2-6 bond [60]. In such receptor specialization for the human body, the key positions in the genetic patterns for subtype H5 viruses may be amino acids 137/Ala, 155/Thr, 163/Ala, 190/Asp, 193/Arg, 225/Asp, 226/Leu, 228/Ser in the NA protein [61, 62]. In the NA protein of avian viruses, 46/Asp; 74/Ser; 151/Asp; 163/Leu; 204/Met; 369/Ile; 396/Met; 420/Gly may be important amino acid positions for adaptation to mammalian animals [63, 64]. Furthermore, a characteristic second receptor site for sialic residues α2-3 (the so-called second hemadsorption center) was identified in the NA molecule of H5N1 avian influenza viruses, whereas such a site was absent in human influenza viruses, which specifically enhanced the affinity of NA to receptors of the α2-6 type [65]. In H5N1 viruses isolated from mink, this center was not detected in its complete form, which brought these isolates closer to human viruses and increased their affinity for α2-6 sialic receptors [66]. An additional site of increased epidemic potential for humans in avian H5N1 viruses may be a deletion in the NA gene and shortening of the NA stalk, which has been frequently found in both avian H5N1 viruses isolated from humans [67, 68], and in pandemic human influenza viruses [69, 70].
The noted structural and functional changes in NP, PB2, HA, NA proteins are the most significant for adaptation and transition of H5 and H7 viruses from birds to humans. At the same time, adaptive changes during host change can be observed in other viral proteins such as PB1, PA, PA-X, NS1, NEP, M1, etc. [71–76], which can be considered as additional extragenic cofactors that enable viral host switching along with the above-described major regulators of the viral host range.
In conclusion, the described adaptive mutations of avian influenza virus genes are necessary for the formation of two main phenotypic traits of human epidemic virus: (1) the property of maintaining a high level of reproduction of the emerged new viral strain in the human body and (2) preservation of high airborne transmission of the virus in the human population after its transfer from birds to humans. The formation of these virus properties and its epidemic potential for the human population is controlled primarily by four major viral proteins NP, PB2, HA, and NA.
Possible transmission ways of highly pathogenic H5 and H7 viruses from birds to humans
There are 2 possible ways of transmission of highly pathogenic influenza viruses from birds to humans (Fig. 3). The first is direct transmission from birds directly to humans. The second possible pathway is transmission of the virus through an intermediary – an intermediate host, which could be a mammalian animal organism. The first route seems to be difficult, because the still non-adapted avian virus must adapt to a new human organism, acquiring the necessary adaptation mutations directly during the process of host change itself. The available observations of several episodes of epidemic outbreaks of highly pathogenic avian influenza H5N1 and H7N9 are consistent with this position. Such outbreaks that occurred in China (1997), Hong Kong (2005), China (2017), and several other areas of the world, caused by direct viral contamination of humans from birds, were localized and did not develop into epidemics [39]. The non-adapted avian virus was not sufficiently infectious to humans and did not cause human-to-human aerosol transmission.
The second possible way of transmission of H5 and H7 subtypes may be through intermediate hosts – a population of mammalians usually linked to birds by a «predator-prey» chain. In this intermediate organism, which is genetically closer to humans, the avian virus will acquire adaptation to the critical mechanisms of the host organism. Such initial pre-adaptation in the mammalian organism can prepare the avian virus and significantly facilitate its transition into the human population and give rise to the formation of a highly pathogenic influenza virus of subtypes H5 and H7. It is important to note that the virus internal protein apparatus (NP, PB2, PB1, M1, NS1), responsible for virus replication, pre-adapted to humans in mammalian organism can give rise not only to the epidemic of highly pathogenic virus of subtypes H5 and/or H7, but also contribute to the emergence of a new antigenic type of epidemic in humans caused by low pathogenic viruses of these subtypes.
Preconditions for the emergence of a pandemic of influenza A viruses
The epidemic situation of the last 2–3 years is characterized with a sharp increase in the incidence of avian and mammalians morbidity in nature and in agriculture with highly pathogenic strain of influenza virus H5N1, belonging to the clade h2.3.4.4b [77–82]. The reported increase in incidence has three features.
- This rise in incidence is associated with the emergence of genetic clade h2.3.4.4.4b of the highly pathogenic subtype of the avian H5N1 virus.
- The spread of this strain among domestic and wild birds is planetary and affects all continents. In the last 2 years, the cumulative mortality of birds and animals from this virus on the planet is approaching 1 billion cases [79, 83].
Fig. 4. Reasons and consequences of the penetration of the highly pathogenic influenza virus A/H5N1 into Northern Eurasia (spring 2005 ‒ spring 2008) [5].
Рис. 4. Причины и последствия проникновения высоковирулентного вируса гриппа A/H5N1 в Северную Евразию (весна 2005 ‒ весна 2008) [5].
The H5 subtype virus appeared in the PRC in February 2003, at the beginning of the spring migration season. It is a reassortant of wild bird influenza A viruses. In 2005, the virus was introduced into the territory of Russia by wild birds with the formation of natural foci (Fig. 4) [5, 84–86]. In recent years in the Russian Federation, outbreaks associated with infection of wild and domestic birds with A/H5N8, A/H5N1, and A/H5N5 viruses have been detected in North Ossetia – Alania, Krasnodar Krai, Astrakhan and Tomsk regions. In 2020, during outbreaks of H5N8 avian influenza in the Astrakhan region the first cases of human infection with this virus were detected – 7 employees were infected at a poultry farm. According to Rosselkhoznadzor, 54 outbreaks of avian influenza have been registered in the Russian Federation in 2022, of which 5 are associated with wild birds, 42 with domestic birds, and 7 with poultry farms. Infection of birds was detected in Magadan, Sakhalin, Khabarovsk Krai, Kaluga, Ivanovo, Orel, Kursk, Belgorod, Samara, Chelyabinsk regions.
- The rise in birds morbidity with the highly pathogenic avian H5N1 virus was accompanied by an increase in the frequency and magnitude of outbreaks of avian influenza in various mammalian species: sea lions, seals, mink, cats, foxes, bears, Arctic foxes and other animal species (a total of 26 mammalian species [87, 88]). The most significant episodes of H5N1 outbreaks have been reported in seals in South and North America [89], mink in Spain [90], cats in Poland [91], fur-bearing animals in Finland [92], and sea lions in Peru [93]. Clinically, the outbreaks were accompanied by generalized multi-organ infection and high mortality, with outbreaks involving tens of thousands of cases (Table 4).
Table 4. Fatal cases among mammals during outbreaks of highly pathogenic avian influenza H5N1 occurred in 2021‒2023
Таблица 4. Смертельные случаи среди млекопитающих при вспышках высокопатогенного птичьего гриппа H5N1 в 2021‒2023 гг.
Regions in the world Регионы в мире | Species of dead animals Вид погибших животных | Number of dead animals Число погибших животных |
North America Северная Америка | Seals Тюлени | > 121 |
South America Южная Америка | Sea lions Морские львы | > 634 |
Spain Испания | Minks Норки | 51 986 |
Poland Польша | Cats Кошки | > 98 |
Finland Финляндия | Fur-bearing farm animals Пушные с/х животные | > 200 000 |
Note. In the table, the data of references [84‒89] are summarized.
Примечание. В таблице суммированы данные публикаций [84‒89].
In the above epizootics, animal-to-animal transmission of the virus was registered, indicating increased contagiousness of the H5N1 virus for mink [87, 90, 92]. This property of the virus is formed both by genes that ensure a sufficient level of virus replication in a given animal (or human) species and by genes that ensure receptor-binding specificity to cell receptors and stability of viral particles to factors of the external and intracellular environment. The most important role in the acquisition of virion resistance to such factors as increased temperature and pH of the external and intracellular environment is played by the conformation of the viral HA protein and especially by the structure of its stem domain [94–98]. Isolated strains of avian H5N1 virus from seals and cats were characterized by the appearance of a number of remarkable mutations in NP, PB2, HA, and NA genes. The detected mutations reflected partial adaptation of the original avian virus to the mammals and were similar to genetic characteristics of human influenza viruses. In particular, the most characteristic mutations E627K and A701K were detected in the polymerase PB2 protein; in the NP protein – G16D, V/I100R, V/Y313F, K357Q; in the HA protein, a linked double mutation 163Ala and 193Arg was detected, which is specific for viral HA having affinity for sialic receptors with CK-α2-6-Gal binding type [38].
In the NA neuraminidase gene of H5N1 viruses isolated during the Spanish mink outbreak in 2021, mutations Ser369Ile and Ile396Met were detected in the second additional receptor binding site with type CKα2-3 (the so-called hemadsorbing center No. 2), inactivating this receptor center [65]. The presence of receptor center No. 2 is characteristic only of avian influenza viruses and is absent in human influenza virus. It is quite obvious that the loss of this site in the NA protein decreases affinity to sialic receptors CKα2-3 and increases affinity for CKα2-6 receptors, which is characteristic of human viruses [38, 66]. The above data show that the highly pathogenic H5N1 influenza viruses that cause disease in mammals have acquired a number of adaptive mutations that distinguish these strains from avian viruses and bring them closer to the human influenza virus. Thus, the H5N1 avian influenza virus has a mixed (avian-human) patterns of mutations showing its partial pre-adaptation to humans. Such ongoing pre-adaptation of avian virus in mammals creates obvious prerequisites for its subsequent advanced transition from mammals to humans. This possibility has been experimentally demonstrated [95, 96].
As noted above, in addition to adaptive mutations, the transition of an avian virus from one host species to another can take place by shifting, i.e., by exchanging genomic RNA segments with viruses already adapted to humans. Such a mechanism of emergence of pandemic influenza A viruses occurred previously in 1957, 1968, and 2009 during the emergence and development of epidemics caused by H2N2, H3N2, and H1N1/09pdm viruses, respectively [11, 12]. The increased frequency of H5N1 avian influenza outbreaks in mammals in 2020–2022 develops preconditions for the realization of a similar RNA reassortment process for this highly pathogenic H5N1 subtype, given the intensive virus genomes exchange and the large number of asymptomatic forms of influenza infection in humans in contact with animals [99, 100]. The transfer of this highly pathogenic H5N1 subtype to mammals increases the probability of co-infection of these animals with avian and human influenza viruses and the subsequent exchange between them of RNA segments responsible for the tropism of the virus to humans, thus creating a real possibility of the emergence of a chimeric highly pathogenic virus that would be able to cross the inter-taxon barrier from birds to humans.
Globalization of the spread of avian influenza
Since the beginning of the 21st century, a wave-like increase in the incidence and spread of HPAI viruses among both domestic and wild birds has been noted [79]. Our monitoring in Northern Eurasia revealed the existence of natural foci associated with aquatic and near-aquatic biocenoses of 15 from the 18 world’s known HA antigenic subtypes of influenza A, including A/H5N1, which was associated with an extensive epizootic and then panzootic among wild and domestic birds in 2003 [19–21, 101–103]. In April 2005, during the spring migration of birds, the H5N1 virus entered the territory of Western Siberia and further spread westward. In April 2008, another genetic lineage of the virus entered the territory of Primorsky Krai with further coverage of more northern territories of the Far East (Fig. 4) [5]. The natural foci of different variants of A/H5N1 and other avian influenza viruses that have emerged in vast areas of northern Eurasia continue to function actively at present. The current paths of spread of H5 subtype HPAI virus strains (clade h.2.3.4.4.4b) also coincide with the migration routes of birds that have benn previously established [83, 88, 101, 104]. These observations support the general concept of the formation of persistent natural foci of the low pathogenic H5 virus, along with other antigenic subtypes of influenza A virus, in Southeast Asia and the Siberian and Far Eastern regions of Russia, which serve as a natural pool and source for the emergence of highly pathogenic strains and their subsequent intra- and intercontinental spread, creating a permanent threat of dangerous pandemics among mammals and humans (shown in Fig. 4). This concept has evolved from extensive basic research on the ecology of viral populations [8, 19–21, 26–29, 101–103].
As mentioned above, the period 2021-2023 saw a major upsurge in avian influenza and the global spread of the highly pathogenic H5N1 virus (clade h2.3.4.4.4b) among birds on all continents of the planet [78–82, 88]. As genetic analysis of viral genomes shows [105], the established reciprocal exchange of highly pathogenic H5N1 virus between mammals and birds in nature has led to the spread and accumulation in nature of viral variants with signs of partial adaptation to mammals, including humans. Also it is a matter of concern and scrutiny that the global rise in avian morbidity is accompanied by increasing outbreaks of H5N1 subtype HPAI among mammals. In the period 2021–2023, 5 large-scale epizootics on 3 continents have been registered among seals in North America, mink in Spain, cats in Poland, fur farm animals in Finland, and sea lions in Peru [87–93]. The magnitude of the animal virus outbreaks is summarized in Table 4. H5N1 influenza viruses isolated from animals in these outbreaks had some of the genetic markers and phenotypic features of high virulence virus characteristics for human and mammalian hosts. Signs of high pathogenicity of avian H5N1 viruses for mammals were manifested in the severe clinical course of the disease, which was accompanied by high mortality, reaching 50–60% of lethality, development of primary viral pneumonia, fatal symptoms of total hemorrhagic inflammation of the lungs (also known as pulmonary hepatization), nervous system damage with severe motor dysfunctions and paralysis, massive hemorrhagic intestinal damage, severe general intoxication, etc. [90, 92, 93].
The observed widespread distribution of such a partially human-adapted H5N1 subtype virus among mammals in close contact with humans poses a real threat of its facilitated transfer to humans with epidemic potential through the respiratory route of transmission among humans. In addition, such animal-adapted avian H5N1 virus can rapidly strengthen and definitively establish its epidemic potential and infectivity through reassortment of genomic RNA segments (mainly NP, PB2, HA, NA) with human influenza viruses, either already during transfer to humans or while still in mammals and birds that have close contact with humans (pigs, ducks and other farm and domestic animals). In pigs and ducks, infection with the highly pathogenic avian H5N1 virus can occur in a subclinical form without pronounced symptoms, which may favor the occurrence of mixed infection with human and avian viruses and the realization of the reassortment process of their genomes [106–108]. According to Avian Influenza Weekly Update No. 919 of October 03, 2023, from January 2003 to the present, 878 cases of human infection with H5N1 virus with 50% lethality, 1568 cases of infection with A/H7N9 virus with 39% lethality, 88 and 90 cases of infection with A/H5N6 and A/H9N2, respectively, with 39% lethality, and single cases of infection caused by viruses of subtypes A/H13N8, A/H7N4, A/H10N3 have been registered in 23 countries [88].
Preventive measures against the threat of avian influenza
The increasing threat for humanity posed by a sharp rise in the incidence and spread of the HPAI virus among animals has brought increased attention to this problem. In this regard, three organizations: The Food and Agriculture Organization of the United Nations (FAO), The World Health Organization (WHO) and The World Organization for Animal Health (WOAH) issued a joint statement which states that «ongoing outbreaks of avian influenza in animals pose a risk to the human population» [87, 88]. This statement set out a common agenda of issues requiring international coordination by government agencies in anticipation and preparation for a possible dangerous avian influenza epidemic. It especially recommended drawing attention to HPAI as a source of risk to human health, strengthening and regulating preventive measures, including biosecurity measures on farms and in the poultry value chain, conducting epidemiological investigations of outbreaks, ensuring rapid transmission and exchange of virus genome data and promoting cooperation between the animal and human health sectors.
Concretizing the preventive measures and necessary preparation in the context of a large-scale threat of avian influenza epidemic, the following main directions of action are proposed:
The proposed measures will make it possible to predict the beginning of dangerous pandemics and their risks, and to minimize the consequences in the event of epidemics caused by avian influenza viruses.
Conclusion
At the present stage, there has been a global upsurge in the infection and morbidity of wild and domestic birds with the H5N1 HPAI virus (clade h2.3.4.4b) and other influenza A viruses. Against this background, mammalian outbreaks caused by this virus clade have become more frequent and highly pathogenic H5N1 and a number of other avian influenza viruses with signs of partial adaptation to humans continue to circulate in nature. Transition of the highly pathogenic H5N1 virus to mammals creates a real threat of its further adaptation to the human body and increase of its epidemic potential with subsequent formation of a pandemic situation. The current pre-epidemic situation necessitates the development of a strategy of preventive measures to minimize the consequences of a possible threatening scenario of pandemic emergence. In order to reduce the risks and consequences of such a scenario, the article identifies urgent measures, including the formation of the effective host-targeted therapy concept.
_______
*The original phrase «For Whom the Bell Tolls» is a title of the novel by Ernest Hemingway.
Note: The work was presented at the international Congress «Molecular Diagnostics and Biosafety – 2024» (Moscow, April 16–17, 2024): Zhirnov O.P., Lvov D.K. Threat of an avian influenza pandemic: formation mechanisms of the population gene pool of the virus. Congress theses, edited by Academician of the Russian Academy of Sciences V.G. Akimkin, Moscow, 2024, pp. 69‒70.
About the authors
Oleg P. Zhirnov
The D.I. Ivaovsky Institute of Virology, The N.F. Gamaleya Research Center of Epidemiology and Microbiology, The Russian Ministry of Health; The Russian-German Academy of Medical-Social and Biotechnological Sciences, Skolkovo Innovation Center
Email: zhirnov@inbox.ru
ORCID iD: 0000-0002-3192-8405
Academician of the Russian Academy of Sciences, Professor, Dr Sci (Biology), Head of the Laboratory
Russian Federation, 123098, Moscow; 109029, MoscowDmitry K. Lvov
The D.I. Ivaovsky Institute of Virology, The N.F. Gamaleya Research Center of Epidemiology and Microbiology, The Russian Ministry of Health
Author for correspondence.
Email: dk_lvov@mail.ru
ORCID iD: 0000-0001-8176-6582
Academician of the Russian Academy of Sciences, Professor, Dr Sci (Medicine), Chief Researcher
Russian Federation, 123098, MoscowReferences
- Walker P.J., Siddell S.G., Lefkowitz E.J., Mushegian A.R., Adriaenssens E.M., Alfenas-Zerbini P., et al. Recent changes to virus taxonomy ratified by the International Committee on Taxonomy of Viruses (2022). Arch. Virol. 2022; 167(11): 2429–40. https://doi.org/10.1007/s00705-022-05516-5
- Klemm C., Boergeling Y., Ludwig S., Ehrhardt C. Immunomodulatory Nonstructural Proteins of Influenza A Viruses. Trends. Microbiol. 2018; 26(7): 624–36. https://doi.org/10.1016/j.tim.2017.12.006
- Abdelwhab E.M., Mettenleiter T.C. Zoonotic Animal Influenza Virus and Potential Mixing Vessel Hosts. Viruses. 2023; 15(4): 980. https://doi.org/10.3390/v15040980
- Sanjuán R., Domingo-Calap P. Mechanisms of viral mutation. Cell Mol. Life Sci. 2016; 73(23): 4433–48. https://doi.org/10.1007/s00018-016-2299-6
- L’vov D.K., Gulyukin M.Yu., Zaberezhnyy A.D., Gulyukin A.M. Formation of population gene pools of zoonotic viruses, potentially threatening biosafety. Voprosy virusologii. 2020; 65(5): 243–58. https://doi.org/10.36233/0507-4088-2020-65-5-1 https://elibrary.ru/kprmam (in Russian)
- Brüssow H. The beginning and ending of a respiratory viral pandemic-lessons from the Spanish flu. Microb. Biotechnol. 2022; 15(5): 1301–17. https://doi.org/10.1111/1751-7915.14053
- Romero-Tejeda A., Capua I. Virus-specific factors associated with zoonotic and pandemic potential. Influenza Other Respir. Viruses. 2013; 7(Suppl. 2): 4–14. https://doi.org/10.1111/irv.12075
- Lvov D.K. Circulation of Influenza viruses in natural biocenosis. In: Viruses and Environment. Academic Press; 1978; 18: 351–80.
- Lvov D.K., Zhdanov V.M. Circulation of influenza viruses genes in the biosphere. Sov. Med. Rev. Virol. 1987; (1): 129–52.
- Lvov D.K. Influenza A viruses – a sum of populations with a common protected gene pool. Sov. Med. Rev. Virol. 1987; (2): 15–37.
- Palese P., Shaw M.L. Orthomyxoviridae: the viruses and their replication. In: Knipe D.M., Howley P.M., eds. Fields Virology. Lippincott Williams & Wilkins; 2007: 1648–89.
- Wille M., Holmes E.C. The ecology and evolution of influenza viruses. Cold Spring Harb. Perspect. Med. 2020; 10(7): a038489. https://doi.org/10.1101/cshperspect.a038489
- Suarez D.L. Evolution of avian influenza viruses. Vet. Microbiol. 2000; 74(1-2): 15–27. https://doi.org/10.1016/s0378-1135(00)00161-9
- Swayne D.E. Changing face of avian influenza ecology and its control : from wild birds to poultry and back again. In: Abstract Book of the 15th World Veterinary Poultry Congress. Beiging; 2007: 98–104.
- Krammer F., Smith G.J.D., Fouchier R.A.M., Peiris M., Kedzierska K., Doherty P.C., et al. Influenza. Nat. Rev. Dis. Primers. 2018; 4(1): 3. https://doi.org/10.1038/s41572-018-0002-y
- Garten W., Klenk H.D. Cleavage activation of the influenza virus hemagglutinin and its role in pathogenesis. In: Klenk H.D., Matrosovich M.N., Stech J., eds. Avian Influenza. Basel: Karger; 2008: 156–67.
- L’vov D.K., Il’ichev V.D. Migrations of Birds and Transfer of Infectious Agents [Migratsii ptits i perenos vozbuditeley infektsiy]. Moscow: Nauka; 1979. (in Russian)
- L’vov D.K., Zhdanov V.M. Persistence of genes of epidemic influenza A viruses in natural populations. Uspekhi sovremennoy biologii. 1982; 93(3): 323–37. (in Russian)
- L’vov D.K., Shchelkanov M.Yu. Avian influenza H5N1. In: L’vov D.K., ed. Viruses and Viral Infections of Humans and Animals. Handbook of Virology [Virusy i virusnye infektsii cheloveka i zhivotnykh. Rukovodstvo po virusologii]. Moscow: MIA; 2013: 554–77. https://elibrary.ru/tlzmhf (in Russian)
- L’vov D.K., Shchelkanov M.Yu., Aliper T.I. Flu of wild birds. In: L’vov D.K ed. Viruses and Viral Infections of Humans and Animals. Handbook of Virology [Virusy i virusnye infektsii cheloveka i zhivotnykh. Rukovodstvo po virusologii]. Moscow: MIA; 2013: 1086–94. (in Russian)
- Lvov D.K., Kaverin N.V. Avian influenza in Northern Eurasia. In: Klenk H.D., Matrosovich M.N., eds. Avian Influenza. Basel: Karger; 2008: 41–58.
- L’vov D.K. Possible significance of natural biocenoses in the variability of influenza A viruses. Voprosy virusologii. 1974; 19(6): 740–4. (in Russian)
- Stallknect D., Brown J.D. Ecology of avian influenza in wild birds. In: Swayne D.E., ed. Avian Influenza. Oxford: Blackwell Publ.; 2008: 43–8.
- Pereira H.G., Tůmová B., Law V.G. Avian influenza A viruses. Bull. World Health Organ. 1965; 32(6): 855–60.
- Becker W.B. The isolation and classification of Tern virus: influenza A-Tern South Africa – 1961. J. Hyg. (Lond.). 1966; 64(3): 309–20. https://doi.org/10.1017/s0022172400040596
- Laver W.G., Webster R.G. Ecology of influenza viruses in lower mammals and birds. Br. Med. Bull. 1979; 35(1): 29–33. https://doi.org/10.1093/oxfordjournals.bmb.a071537
- Webster R.G., Laver W.G. Further evidence to support a recommendational events in the origin of new pandemic influenza viruses. In: Kilbourne E., ed. The Influenza. NY-San-Francisco: Academic Press; 1975; (7): 299–308.
- L’vov D.K. Population interactions in biological system: influenza virus a – wild and domestic animals- human; reasons and consequences of introduction high pathogenic influenza virus A/H5N1 on Russian territory. Zhurnal mikrobiologii epidemiologii i immunobiologii. 2006; 93(3): 96–100. https://elibrary.ru/htqbwt (in Russian)
- L’vov D.K., Al’khovskiy S.D. To the 55th anniversary of the Department of Virus Ecology with the Scientific and Practical Center for the Ecology and Epidemiology of Influenza (D.I. Ivanovsky Institute of Virology of the N.F. Gamaleya National Research Center for Epidemiology and Microbiology of the Ministry of Health of Russian Federation). Voprosy virusologii. 2024; 69(1): 7–21. https://doi.org/10.36233/0507-4088-217 https://elibrary.ru/xdikxk (in Russian)
- Böttcher-Friebertshäuser E., Garten W., Matrosovich M., Klenk H.D. The hemagglutinin: a determinant of pathogenicity. Curr. Top. Microbiol. Immunol. 2014; 385: 3–34. https://doi.org/10.1007/82_2014_384
- Klenk H.D., Rott R., Orlich M., Blödorn J. Activation of influenza A viruses by trypsin treatment. Virology. 1975; 68(2): 426–39. https://doi.org/10.1016/0042-6822(75)90284-6
- Lazarowitz S.G., Choppin P.W. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology. 1975; 68(2): 440–54. https://doi.org/10.1016/0042-6822(75)90285-8
- Zhirnov O.P. The phenomenon of proteolytic activation of myxoviruses and a new strategy for treating viral diseases. Voprosy virusologii. 1983; 28(4): 9–21. (Russian)
- Zhirnov O.P., Klenk H.D., Wright P.F. Aprotinin and similar protease inhibitors as drugs against influenza. Antiviral. Res. 2011; 92(1): 27–36. https://doi.org/10.1016/j.antiviral.2011.07.014
- Zhirnov O.P., Ikizler M.R., Wright P.F. Cleavage of influenza a virus hemagglutinin in human respiratory epithelium is cell associated and sensitive to exogenous antiproteases. J. Virol. 2002; 76(17): 8682–9. https://doi.org/10.1128/jvi.76.17.8682-8689.2002
- Zhirnov O.P., Matrosovich T.Y., Matrosovich M.N., Klenk H.D. Aprotinin, a protease inhibitor, suppresses proteolytic activation of pandemic H1N1v influenza virus. Antivir. Chem. Chemother. 2011; 21(4): 169–74. https://doi.org/10.3851/IMP1715
- Garten W., Hallenberger S., Ortmann D., Schäfer W., Vey M., Angliker H., et al. Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie. 1994; 76(3-4): 217–25. https://doi.org/10.1016/0300-9084(94)90149-x
- Chauhan R.P., Gordon M.L. An overview of influenza A virus genes, protein functions, and replication cycle highlighting important updates. Virus Genes. 2022; 58(4): 255–69. https://doi.org/10.1007/s11262-022-01904-w
- Lai S., Qin Y., Cowling B.J., Ren X., Wardrop N.A., Gilbert M., et al. Global epidemiology of avian influenza A H5N1 virus infection in humans, 1997–2015: a systematic review of individual case data. Lancet Infect. Dis. 2016; 16(7): e108–18. https://doi.org/10.1016/S1473-3099(16)00153-5
- Haller O., Kochs G. Mx genes: host determinants controlling influenza virus infection and trans-species transmission. Hum. Genet. 2020; 139(6–7): 695–705. https://doi.org/10.1007/s00439-019-02092-8
- Turan K., Mibayashi M., Sugiyama K., Saito S., Numajiri A., Nagata K. Nuclear MxA proteins form a complex with influenza virus NP and inhibit the transcription of the engineered influenza virus genome. Nucleic. Acids Res. 2004; 32(2): 643–52. https://doi.org/10.1093/nar/gkh192
- Peacock T.P., Sheppard C.M., Lister M.G., Staller E., Frise R., Swann O.C., et al. Mammalian ANP32A and ANP32B proteins drive differential polymerase adaptations in avian influenza virus. J. Virol. 2023; 97(5): e0021323. https://doi.org/10.1128/jvi.00213-23
- Sheppard C.M., Goldhill D.H., Swann O.C., Staller E., Penn R., Platt O.K., et al. An influenza A virus can evolve to use human ANP32E through altering polymerase dimerization. Nat. Commun. 2023; 14(1): 6135. https://doi.org/10.1038/s41467-023-41308-4
- Tome-Amat J., Ramos I., Amanor F., Fernández-Sesma A., Ashour J. Influenza A virus utilizes low-affinity, high-avidity interactions with the nuclear import machinery to ensure infection and immune evasion. J. Virol. 2018; 93(1): e01046–18. https://doi.org/10.1128/JVI.01046-18
- Zhang B., Xu S., Liu M., Wei Y., Wang Q., Shen W., et al. The nucleoprotein of influenza A virus inhibits the innate immune response by inducing mitophagy. Autophagy. 2023; 19(7): 1916–33. https://doi.org/10.1080/15548627.2022.2162798
- Ninpan K., Suptawiwat O., Boonarkart C., Phuangphung P., Sathirareuangchai S., Uiprasertkul M., et al. Expression of importin-α isoforms in human nasal mucosa: implication for adaptation of avian influenza A viruses to human host. Virol. J. 2016; 13: 90. https://doi.org/10.1186/s12985-016-0546-y
- Morris A.K., Wang Z., Ivey A.L., Xie Y., Hill P.S., Schey K.L., et al. Cellular mRNA export factor UAP56 recognizes nucleic acid binding site of influenza virus NP protein. Biochem. Biophys. Res. Commun. 2020; 525(2): 259–64. https://doi.org/10.1016/j.bbrc.2020.02.059
- Mänz B., Dornfeld D., Götz V., Zell R., Zimmermann P., Haller O., et al. Pandemic influenza A viruses escape from restriction by human MxA through adaptive mutations in the nucleoprotein. PLoS Pathog. 2013; 9(3): e1003279. https://doi.org/10.1371/journal.ppat.1003279
- Zhirnov O.P. The host origin of influenza A viruses can be assessed by the intracellular cleavage of the viral nucleocapsid protein. Brief report. Arch. Virol. 1988; 99(3-4): 277–84. https://doi.org/10.1007/BF01311077
- Zhirnov O., Bukrinskaya A.G. Nucleoproteins of animal influenza viruses, in contrast to those of human strains, are not cleaved in infected cells. J. Gen. Virol. 1984; 65 (Pt. 6): 1127–34. https://doi.org/10.1099/0022-1317-65-6-1127
- Finkelstein D.B., Mukatira S., Mehta P.K., Obenauer J.C., Su X., Webster R.G., et al. Persistent host markers in pandemic and H5N1 influenza viruses. J. Virol. 2007; 81(19): 10292–9. https://doi.org/10.1128/JVI.00921-07
- Worobey M., Han G.Z., Rambaut A. Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc. Natl. Acad. Sci. USA. 2014; 111(22): 8107–12. https://doi.org/10.1073/pnas.1324197111
- Chen G.W., Gong Y.N., Shih S.R. Influenza A virus plasticity-A temporal analysis of species-associated genomic signatures. J. Formos. Med. Assoc. 2015; 114(5): 456–63. https://doi.org/10.1016/j.jfma.2015.01.015
- Long J.S., Idoko-Akoh A., Mistry B., Goldhill D., Staller E., Schreyer J., et al. Species-specific differences in use of ANP32 proteins by influenza A virus. Elife. 2019; 8: e45066. https://doi.org/10.7554/eLife.45066
- Subbarao E.K., London W., Murphy B.R. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 1993; 67(4): 1761–4. https://doi.org/10.1128/jvi.67.4.1761-1764.1993
- Mitnaul L.J., Matrosovich M.N., Castrucci M.R., Tuzikov A.B., Bovin N.V., Kobasa D., et al. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J. Virol. 2000; 74(13): 6015–20. https://doi.org/10.1128/jvi.74.13.6015-6020.2000
- Kaverin N.V., Matrosovich M.N., Gambaryan A.S., Rudneva I.A., Shilov A.A., Varich N.L., et al. Intergenic HA-NA interactions in influenza A virus: postreassortment substitutions of charged amino acid in the hemagglutinin of different subtypes. Virus Res. 2000; 66(2): 123–9. https://doi.org/10.1016/s0168-1702(99)00131-8
- Wagner R., Matrosovich M., Klenk H.D. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev. Med. Virol. 2002; 12(3): 159–66. https://doi.org/10.1002/rmv.352
- Gambaryan A.S., Matrosovich M.N. What adaptive changes in hemagglutinin and neuraminidase are necessary for emergence of pandemic influenza virus from its avian precursor? Biokhimiya. 2015; 80(7): 872–80. https://doi.org/10.1134/S000629791507007X (in Russian)
- Eggink D., Spronken M., van der Woude R., Buzink J., Broszeit F., McBride R., et al. Phenotypic effects of substitutions within the receptor binding site of highly pathogenic avian influenza H5N1 virus observed during human infection. J. Virol. 2020; 94(13): e00195-20. https://doi.org/10.1128/JVI.00195-20
- Guo H., de Vries E., McBride R., Dekkers J., Peng W., Bouwman K.M., et al. Highly pathogenic influenza A (H5Nx) viruses with altered H5 receptor-binding specificity. Emerg. Infect. Dis. 2017; 23(2): 220–31. https://doi.org/10.3201/eid2302.161072
- Gao R., Gu M., Liu K., Li Q., Li J., Shi L., et al. T160A mutation-induced deglycosylation at site 158 in hemagglutinin is a critical determinant of the dual receptor binding properties of clade 2.3.4.4 H5NX subtype avian influenza viruses. Vet. Microbiol. 2018; 217: 158–66. https://doi.org/10.1016/j.vetmic.2018.03.018
- Leguia M., Garcia-Glaessner A., Muñoz-Saavedra B., Juarez D., Barrera P., Calvo-Mac C., et al. Highly pathogenic avian influenza A (H5N1) in marine mammals and seabirds in Peru. Nat. Commun. 2023; 14(1): 5489. https://doi.org/10.1038/s41467-023-41182-0
- Scheibner D., Salaheldin A.H., Bagato O., Zaeck L.M., Mostafa A., Blohm U., et al. Phenotypic effects of mutations observed in the neuraminidase of human origin H5N1 influenza A viruses. PLoS Pathog. 2023; 19(2): e1011135. https://doi.org/10.1371/journal.ppat.1011135
- Du W., de Vries E., van Kuppeveld F.J.M., Matrosovich M., de Haan C.A.M. Second sialic acid-binding site of influenza A virus neuraminidase: binding receptors for efficient release. FEBS J. 2021; 288(19): 5598–612. https://doi.org/10.1111/febs.15668
- de Vries E., de Haan C.A. Letter to the editor: Highly pathogenic influenza A(H5N1) viruses in farmed mink outbreak contain a disrupted second sialic acid binding site in neuraminidase, similar to human influenza A viruses. Euro Surveill. 2023; 28(7): 2300085. https://doi.org/10.2807/1560-7917.ES.2023.28.7.2300085
- Bender C., Hall H., Huang J., Klimov A., Cox N., Hay A., et al. Characterization of the surface proteins of influenza A (H5N1) viruses isolated from humans in 1997-1998. Virology. 1999; 254(1): 115–23. https://doi.org/10.1006/viro.1998.9529
- Zhou H., Yu Z., Hu Y., Tu J., Zou W., Peng Y., et al. The special neuraminidase stalk-motif responsible for increased virulence and pathogenesis of H5N1 influenza A virus. PLoS One. 2009; 4(7): e6277. https://doi.org/10.1371/journal.pone.0006277
- Zhirnov O.P., Vorobjeva I.V., Saphonova O.A., Poyarkov S.V., Ovcharenko A.V., Anhlan D., et al. Structural and evolutionary characteristics of HA, NA, NS and M genes of clinical influenza A/H3N2 viruses passaged in human and canine cells. J. Clin. Virol. 2009; 45(4): 322–33. https://doi.org/10.1016/j.jcv.2009.05.030
- Young S.G., Kitchen A., Kayali G., Carrel M. Unlocking pandemic potential: prevalence and spatial patterns of key substitutions in avian influenza H5N1 in Egyptian isolates. BMC Infect. Dis. 2018; 18(1): 314. https://doi.org/10.1186/s12879-018-3222-6
- Nogales A., Villamayor L., Utrilla-Trigo S., Ortego J., Martinez-Sobrido L., DeDiego M.L. Natural selection of H5N1 avian influenza A viruses with increased PA-X and NS1 shutoff activity. Viruses. 2021; 13(9): 1760. https://doi.org/10.3390/v13091760
- Taft A.S., Ozawa M., Fitch A., Depasse J.V., Halfmann P.J., Hill-Batorski L., et al. Identification of mammalian-adapting mutations in the polymerase complex of an avian H5N1 influenza virus. Nat. Commun. 2015; 6: 7491. https://doi.org/10.1038/ncomms8491
- Elgendy E.M., Arai Y., Kawashita N., Daidoji T., Takagi T., Ibrahim M.S., et al. Identification of polymerase gene mutations that affect viral replication in H5N1 influenza viruses isolated from pigeons. J. Gen. Virol. 2017; 98(1): 6–17. https://doi.org/10.1099/jgv.0.000674
- Wang C., Qu R., Zong Y., Qin C., Liu L., Gao X., et al. Enhanced stability of M1 protein mediated by a phospho-resistant mutation promotes the replication of prevailing avian influenza virus in mammals. PLoS Pathog. 2022; 18(7): e1010645. https://doi.org/10.1371/journal.ppat.1010645
- Cheung P.H., Lee T.T., Chan C.P., Jin D.Y. Influenza A virus PB1-F2 protein: An ambivalent innate immune modulator and virulence factor. J. Leukoc. Biol. 2020; 107(5): 763–71. https://doi.org/10.1002/JLB.4MR0320-206R
- Rashid F., Xie Z., Li M., Xie Z., Luo S., Xie L. Roles and functions of IAV proteins in host immune evasion. Front. Immunol. 2023; 14: 1323560. https://doi.org/10.3389/fimmu.2023.1323560
- WHO. Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003–2020. Available at: https://www.who.int/influenza/human_animal_interface/2020_10_07_tableH5N1.pdf
- Lewis N.S., Banyard A.C., Whittard E., Karibayev T., Al Kafagi T., Chvala I., et al. Emergence and spread of novel H5N8, H5N5 and H5N1 clade 2.3.4.4 highly pathogenic avian influenza in 2020. Emerg. Microbes. Infect. 2021; 10(1): 148–51. https://doi.org/10.1080/22221751.2021.1872355
- Shi J., Zeng X., Cui P., Yan C., Chen H. Alarming situation of emerging H5 and H7 avian influenza and effective control strategies. Emerg. Microbes. Infect. 2023; 12(1): 2155072. https://doi.org/10.1080/22221751.2022.2155072.
- Sobolev I., Sharshov K., Dubovitskiy N., Kurskaya O., Alekseev A., Leonov S., et al. Highly pathogenic avian influenza A(H5N8) virus clade 2.3.4.4b, Western Siberia, Russia, 2020. Emerg. Infect. Dis. 2021; 27(8): 2224–7. https://doi.org/10.3201/eid2708.204969
- Tian J., Bai X., Li M., Zeng X., Xu J., Li P., et al. Highly Pathogenic Avian Influenza Virus (H5N1) Clade 2.3.4.4b Introduced by Wild Birds, China, 2021. Emerg. Infect. Dis. 2023; 29(7): 1367–75. https://doi.org/10.3201/eid2907.221149
- Isoda N., Onuma M., Hiono T., Sobolev I., Lim H.Y., Nabeshima K., et al. Detection of new H5N1 high pathogenicity avian influenza viruses in winter 2021-2022 in the Far East, which are genetically close to those in Europe. Viruses. 2022; 14(10): 2168. https://doi.org/10.3390/v14102168
- Adlhoch C., Fusaro A., Gonzales J.L., Kuiken T., Mirinavičiūtė G., Niqueux É., et al. Avian influenza overview June-September 2023. EFSA J. 2023; 21(10): e08328. https://doi.org/10.2903/j.efsa.2023.8328
- WHO (Western Pacific Ocean Region). Human infection with avian influenza A/H5 viruses. Human infection with avian influenza A(H5N1) virus. Wkly Update Number. 852; 2022.
- WHO (Western Pacific Ocean Region). Human infection with avian influenza A(H5) viruses. Human infection with avian influenza A(H5N1) virus. Avian Influ. Wkly Update Number. 921 (Pt. 1); 2023.
- L’vov D.K., Borisevich S.V., Al’khovskiy S.V., Burtseva E.I. Relevant approaches to analysis of viral genomes for biosafety. Infektsionnye bolezni: novosti, mneniya, obuchenie. 2019; 8(2): 96–101. https://doi.org/10.24411/2305-3496-2019-12012 https://elibrary.ru/xbkmpl (in Russian)
- WHO: Ongoing avian influenza outbreaks in animals pose risk to humans; 2023. Available at: https://who.int/news/item/12-07-2023-ongoing-avian-influenza-outbreaks-in-animals-pose-risk-to-humans
- Adlhoch C., Fusaro A., Gonzales J.L., Kuiken T., Marangon S., Mirinaviciute G., et al. Avian influenza overview December 2022 – March 2023. EFSA J. 2023; 21(3): e07917. https://doi.org/10.2903/j.efsa.2023.7917
- Puryear W., Sawatzki K., Hill N., Foss A., Stone J.J., Doughty L., et al. Highly pathogenic avian influenza A(H5N1) virus outbreak in New England Seals, United States. Emerg. Infect. Dis. 2023; 29(4): 786–91. https://doi.org/10.3201/eid2904.221538
- Agüero M., Monne I., Sánchez A., Zecchin B., Fusaro A., Ruano M.J., et al. Highly pathogenic avian influenza A(H5N1) virus infection in farmed minks, Spain, October 2022. Euro Surveill. 2023; 28(3): 2300001. https://doi.org/10.2807/1560-7917.ES.2023.28.3.2300001
- Rabalski L., Milewska A., Pohlmann A., Gackowska K., Lepionka T., Szczepaniak K., et al. Emergence and potential transmission route of avian influenza A (H5N1) virus in domestic cats in Poland, June 2023. Euro. Surveill. 2023; 28(31): 2300390. https://doi.org/10.2807/1560-7917.ES.2023.28.31.2300390
- Lindh E., Lounela H., Ikonen N., Kantala T., Savolainen-Kopra C., Kauppinen A., et al. Highly pathogenic avian influenza A(H5N1) virus infection on multiple fur farms in the South and Central Ostrobothnia regions of Finland, July 2023. Euro. Surveill. 2023; 28(31): 2300400. https://doi.org/10.2807/1560-7917.ES.2023.28.31.2300400
- Gamarra-Toledo V., Plaza P.I., Gutiérrez R., Inga-Diaz G., Saravia-Guevara P., Pereyra-Meza O., et al. Mass mortality of sea lions caused by highly pathogenic avian influenza A(H5N1) virus. Emerg. Infect. Dis. 2023; 29(12): 2553–6. https://doi.org/10.3201/eid2912.230192
- Russier M., Yang G., Rehg J.E., Wong S.S., Mostafa H.H., Fabrizio T.P., et al. Molecular requirements for a pandemic influenza virus: An acid-stable hemagglutinin protein. Proc. Natl Acad. Sci. USA. 2016; 113(6): 1636–41. https://doi.org/10.1073/pnas.1524384113
- Herfst S., Schrauwen E.J., Linster M., Chutinimitkul S., de Wit E., Munster V.J., et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012; 336(6088): 1534–41. https://doi.org/10.1126/science.1213362
- Imai M., Watanabe T., Hatta M., Das S.C., Ozawa M., Shinya K., et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012; 486(7403): 420–8. https://doi.org/10.1038/nature10831
- Tosheva I.I., Saygan K.S., Mijnhardt S.M., Russell C.J., Fraaij P., Herfst S. Hemagglutinin stability as a key determinant of influenza A virus transmission via air. Curr. Opin. Virol. 2023; 61: 101335. https://doi.org/10.1016/j.coviro.2023.101335
- Richard M., Fouchier R.A. Influenza A virus transmission via respiratory aerosols or droplets as it relates to pandemic potential. FEMS Microbiol. Rev. 2016; 40(1): 68–85. https://doi.org/10.1093/femsre/fuv039
- Furuya-Kanamori L., Cox M., Milinovich G.J., Magalhaes R.J., Mackay I.M., Yakob L. Heterogeneous and dynamic prevalence of asymptomatic influenza virus infections. Emerg. Infect. Dis. 2016; 22(6): 1052–6. https://doi.org/10.3201/eid2206.151080
- Nguyen T.T.K., Ngo T.T., Tran P.M., Pham T.T.T., Vu H.T.T., Nguyen N.T.H., et al. Respiratory viruses in individuals with a high frequency of animal exposure in southern and highland Vietnam. J. Med. Virol. 2020; 92(8): 971–81. https://doi.org/10.1002/jmv.25640
- Lvov D.K., Shchelkanov M.Y., Alkhovsky S.V., Deryabin P.G. Zoonotic Viruses of northern Eurasia: Taxonomy and Ecology. London: Academic Press, Elsevier; 2015.
- Lvov D.K., Shchelkanov M.Y., Prilipov A.G., Vlasov N.A., Fedyakina I.T., Deryabin P.G., et al. Evolution of highly pathogenic avian influenza H5N1 virus in natural ecosystems of northern Eurasia (2005-08). Avian. Dis. 2010; 54(1 Suppl.): 483–95. https://doi.org/10.1637/8893-042509-Review.1
- Alexander D.J. Summary of avian influenza activity in Europe, Asia, Africa, and Australasia, 2002–2006. Avian. Dis. 2007; 51(1 Suppl.): 161–6. https://doi.org/10.1637/7602-041306R.1
- Yang Q., Wang B., Lemey P., Dong L., Mu T., Wiebe R.A., et al. Synchrony of bird migration with avian influenza global spread; implications for vulnerable bird orders. bioRxiv. Preprint. https://doi.org/10.1101/2023.05.22.541648
- Huang P., Sun L., Li J., Wu Q., Rezaei N., Jiang S., et al. Potential cross-species transmission of highly pathogenic avian influenza H5 subtype (HPAI H5) viruses to humans calls for the development of H5-specific and universal influenza vaccines. Cell Discov. 2023; 9(1): 58. https://doi.org/10.1038/s41421-023-00571-x
- Soda K., Tomioka Y., Usui T., Ozaki H., Ito H., Nagai Y., et al. Susceptibility of common dabbling and diving duck species to clade 2.3.2.1 H5N1 high pathogenicity avian influenza virus: an experimental infection study. J. Vet. Med. Sci. 2023; 85(9): 942–9. https://doi.org/10.1292/jvms.23-0122
- Lee S.H., Lee J., Noh J.Y., Jeong J.H., Kim J.B., Kwon J.H., et al. Age is a determinant factor in the susceptibility of domestic ducks to H5 clade 2.3.2.1c and 2.3.4.4e high pathogenicity avian influenza viruses. Front. Vet. Sci. 2023; 10: 1207289. https://doi.org/10.3389/fvets.2023.1207289
- Graaf A., Piesche R., Sehl-Ewert J., Grund C., Pohlmann A., Beer M., et al. Low susceptibility of pigs against experimental infection with HPAI virus H5N1 Clade h2.3.4.4b. Emerg. Infect. Dis. 2023; 29(7): 1492–5. https://doi.org/10.3201/eid2907.230296
- Subbiah J., Oh J., Kim K.H., Shin C.H., Park B.R., Bhatnagar N., et al. A chimeric thermostable M2e and H3 stalk-based universal influenza A virus vaccine. NPJ Vaccines. 2022; 7(1): 68. https://doi.org/10.1038/s41541-022-00498-6
- He X., Zhang T., Huan S., Yang Y. Novel influenza vaccines: from Research and Development (R&D) challenges to regulatory responses. Vaccines (Basel). 2023; 11(10): 1573. https://doi.org/10.3390/vaccines11101573
- Misplon J.A., Lo C.Y., Crabbs T.A., Price G.E., Epstein S.L. Adenoviral-vectored universal influenza vaccines administered intranasally reduce lung inflammatory responses upon viral challenge 15 months post-vaccination. J. Virol. 2023; 97(10): e0067423. https://doi.org/10.1128/jvi.00674-23
- Tripp R.A. Understanding immunity to influenza: implications for future vaccine development. Expert. Rev. Vaccines. 2023; 22(1): 871–5. https://doi.org/10.1080/14760584.2023.2266033
- WHO. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. Available at: https://who.int/influenza/vaccines/virus/characteristicsvirusvaccines/en
- WHO. Antigenic and genetic characteristics of zoonotic influenza A viruses and development of candidate vaccine viruses for pandemic preparedness in the 2024 southern hemisphere influenza season. Available at: https://cdn.who.int/media/docs/default-source/influenza/who-influenza-recommendations/vcm-southern-hemisphere-recommendation-2024/202309_zoonotic_vaccinvirusupdate.pdf?sfvrsn=e78676a0_5
- Ludwig S., Pleschka S., Planz O. MEK inhibitors as novel host-targeted antivirals with a dual-benefit mode of action against hyperinflammatory respiratory viral diseases. Curr. Opin. Virol. 2023; 59: 101304. https://doi.org/10.1016/j.coviro.2023.101304
- Chakraborty S., Chauhan A. Fighting the flu: a brief review on anti-influenza agents. Biotechnol. Genet. Eng. Rev. 2023; 1–52. https://doi.org/10.1080/02648725.2023.2191081
- Blake M.E., Kleinpeter A.B., Jureka A.S., Petit C.M. Structural Investigations of Interactions between the Influenza a Virus NS1 and Host Cellular Proteins. Viruses. 2023; 15(10): 2063. https://doi.org/10.3390/v15102063
- L’vov D.K., Al’khovskiy S.D., Zhirnov O.P. 130th anniversary of virology. Voprosy virusologii. 2022; 67(5): 357–84. https://doi.org/10.36233/0507-4088-140 https://elibrary.ru/qhembl
Supplementary files