Markers of antroponotic viral infections in vervet monkeys arrived from their natural habitat (Tanzania)
- Authors: Dogadov D.I.1, Kyuregyan K.K.2,3, Alexandra G.M.1, Minosyan A.A.1, Kochkonyan A.A.1, Karlsen A.A.2,3, Vyshemirsky O.I.1, Karal-ogly D.D.1, Mikhailov M.I.2,3
-
Affiliations:
- Research Institute of Medical Primatology of the Ministry of Education and Science of Russia
- Central Research Institute of Epidemiology of the Federal Service for Surveillance on Consumer Rights Protection and Human Wellbeing
- I.I. Mechnikov Research Institute of Vaccines and Sera
- Issue: Vol 68, No 5 (2023)
- Pages: 394-403
- Section: ORIGINAL RESEARCHES
- URL: https://virusjour.crie.ru/jour/article/view/14733
- DOI: https://doi.org/10.36233/0507-4088-188
- EDN: https://elibrary.ru/awajxs
- ID: 14733
Cite item
Abstract
Introduction. Various human viruses have been identified in wild monkeys and in captive primates. Cases of transmission of viruses from wild monkeys to humans and vice versa are known.
The aim of this study was to identify markers of anthroponotic viral infections in vervet monkeys (Chlorocebus pygerythrus) arrived from their natural habitat (Tanzania).
Materials and methods. Fecal samples (n = 56) and blood serum samples (n = 75) obtained from 75 animals, respectively, on days 10 and 23 after admission to the primate center, were tested for the markers of anthroponotic viral infections (Ebola virus, Marburg virus, lymphocytic choriomeningitis, hepatitis C virus, herpes simplex virus (HSV), cytomegalovirus (CMV), Epstein–Barr virus (EBV), parainfluenza types 1 and 3, intestinal adenoviruses, rotaviruses) by enzyme immunoassay (ELISA) and polymerase chain reaction (PCR).
Results and discussion. Among the examined animals, markers of 6 out of 11 tested viral infections were identified. Detection rates of IgG antibodies to HSV-1,2 (15.9%) and CMV (15.9%) were two times as low as IgG antibodies to EBV (31.8%). Among the markers of respiratory viral infections, IgG antibodies to parainfluenza virus type 1 were found (6.8%). 14.3% of the animals had rotavirus antigen, and 94% had simian adenovirus DNA. Markers of hemorrhagic fevers Ebola, Marburg, LCM, hepatitis C, and type 3 parainfluenza were not detected.
Conclusion. When importing monkeys from different regions of the world, an expanded screening for viral infections is needed considering the epidemiological situation both in the country of importation and in the country of destination.
Full Text
Introduction
Viral infections pose a potential threat to the health of wild and laboratory primate populations as well as to the personnel involved in their care. This is particularly true at facilities where there is frequent turnover or movement of animals or at facilities and sites where monkeys imported from natural habitats are introduced into colonies of susceptible animals [1].
Primates, along with rodents and bats, are the most common reservoir and source of zoonotic viral infections compared to other groups of mammals [2]. The close evolutionary relationship between humans and primates facilitates the cross-species transmission of various pathogens [3]. Good examples are human coronavirus OC43 identified in wild chimpanzees in Côte d’Ivoire [4] and SARS-CoV-2 detected in captive gorillas after the exposure to an infected, though asymptomatic staff member of the San Diego Zoo [5]. In addition, a large number of human viruses, including coronaviruses, herpesviruses, rotaviruses, and enteroviruses, viruses causing hepatitis, enteric adenoviruses, have been found in captive and wild primates [6–13]. Many important human pathogens such as the yellow fever virus, Zika virus, dengue virus, and HIV were passed to humans through zoonotic transmission from primates [5, 14, 15]. Conversely, some viruses found in non-human primates, such as poliovirus and measles virus, are believed to be derived from the human population [16–18].
In addition, the likelihood of pathogen transmission increases through public interaction with monkeys at zoos, primate centers, and during wild nature travel being one of the most popular avenues of ecotourism. A convincing example is Bali Island where more than 700,000 tourists annually visit temples inhabited by primates. Researchers described the case of infection of a tourist with the simian foamy virus after the contact with primates in a temple [3].
Based on the current statistics, a total of 140 monkey species are susceptible to infection with 186 DNA and RNA viruses, around 70% of them being also found in humans [5]. In Russia, monkeys of the Sukhumi Primate Center had spontaneous viral infections pathogenic to humans, such as measles, polio, hepatitis A virus (HAV), encephalomyocarditis, seasonal coronavirus infection, and simian hemorrhagic fever [19].
In recent decades, cross-species virus transmission between animals and humans has been a major source of emerging infectious diseases, being a global public health concern. The pandemic caused by SARS-CoV-2, which spread around the world [5], is an illustrative example [5].
Thus, studies of viral diversity in monkeys are of high importance for limitation of potential transmission of viruses between humans and various species of primates.
Previously, in compliance with the quarantine requirements, we examined the green monkeys admitted to the Research Institute of Medical Primatology in June 2014 for the presence of markers of enteric viral hepatitis and respiratory infections (measles virus and adenovirus). Some of the examined animals were tested positive for markers of HAV infection (anti-HAV IgG – 63.1%, anti-HAV IgM – 27.5%, HAV Ag – 27.5%, HAV RNA – 27.5%) and respiratory adenovirus infection (anti-IgG – 14.8%, anti-IgM – 7.4%), while no markers of infection caused by the hepatitis E virus (HEV) and measles infection were detected [6, 10, 20].
The purpose of this study was the further, more extensive detection of serological and molecular genetic markers of anthroponotic viral infections in the vervet monkeys (Chlorocebus pygerythrus), which came from their natural habitats (Tanzania).
Materials and methods
The study was performed using serum and fecal samples from 75 vervet monkeys (Chlorocebus pygerythrus), which came from their natural habitats (Tanzania) in 2014. Fecal samples (n = 56) were collected on the 10th day, and serum samples (n = 75) were collected on the 23rd day after the animals arrived. After they had been collected in 2014, the fecal and serum samples were stored frozen in several aliquots at −70 °C. The tests for viral infection markers were performed both on a real-time basis (enteric adenovirus, Ebola and Marburg viruses, lymphocytic choriomeningitis virus (LCMV), rotavirus) and retrospectively (herpes simplex virus types 1 and 2 (HSV-1, HSV-2), cytomegalovirus (CMV), Epstein-Barr virus (EBV), parainfluenza virus type 1 (PI-1) and type 3 (PI-3), hepatitis C virus (HCV)).
The authors confirm the compliance with institutional and national standards for the use of laboratory animals in accordance with the Consensus Author Guidelines on Animal Ethics and Welfare (IAVES, July 23, 2010). The research protocol was approved by the Ethics Committee (minutes No. 135, 20/5/2014).
Antibodies to HCV (anti-HCV), HSV-1,2 (anti-HSV-1,2), CMV (anti-CMV), and EBV (anti-EBV) were detected using the enzyme-linked immunosorbent assay (ELISA) and commercial ELISA-ANTI-HCV, DS-ELISA-ANTI-HSV-1,2-G, DS-ELISA-ANTI-CMV-G, and DS-ELISA-ANTI-EBV-VCA-G reagent kits (Diagnostic Systems, Russia). ELISA-Parainfluenza-1-IgG and ELISA-Parainfluenza-3-IgG reagent kits were used for detection of IgG antibodies to PI-1 and PI-3 (anti-PI-1 and anti-PI-3) (ECOlab, Russia).
The conjugate to human immunoglobulins from DS-ELISA-ANTI-HSV-1,2-G, DS-ELISA-ANTI-CMV-G, and DS-ELISA-ANTI-EBV-VCA-G, and ELISA-Parainfluenza-1-IgG reagent kits was compared through testing reactive and non-reactive serum panels with the conjugate to monkey immunoglobulins RABBIT ANTI-MONKEY IgG (MERCK, USA) in dilutions ranging from 1 : 2500 to 1 : 200,000, depending on the test. The average optical density values at the wavelength of 450 nm (OD450), which were obtained in tests with both conjugates, were compared using the Mann–Whitney U test.
The detection of the antigen of the group A rotavirus was performed in fecal samples using a commercial ELISA-Rota-Ag reagent kit (Vector Best, Russia).
The ELISA results were read and calculated on an ImmunoChem-2100 spectrophotometer (Intermedica, USA). The calculated results were expressed in OD450 units; additionally, for HSV, OD450 was expressed in titers, and for CMV – in IU/ml.
Nucleic acids were extracted from the 10% fecal suspension using a RIBO-prep kit (InterLabService, Russia) in accordance with the manufacturer’s instruction.
The polymerase chain reaction (PCR) was used to detect adenovirus DNA in fecal samples from monkeys with application of primers for the hexon gene of the most medically important human adenoviruses of groups A–F [21]. The results were analyzed by electrophoresis of PCR products using a reagent kit for electrophoretic detection (InterLabService, Russia). The amplicons were purified from agarose gel using the QIAquick Gel Extraction Kit (Qiagen, Germany) and Sanger sequencing was performed using an automated ABI 3500 genetic analyzer (ABI, USA) and a Big Dye Terminator v. 3.1 reagent kit in accordance with the manufacturer’s protocol.
The derived nucleotide sequences were aligned with each other and with the respective regions of full or partial adenovirus genome sequences available in GenBank at the time of the study using the MEGA X software. The phylogenetic analysis was conducted to confirm the specificity and to identify adenovirus species using the current-year ICTV classification for the genus Mastadenovirus. The phylogenetic tree was constructed using the PhyML 3.3 software based on the ML (maximum likelihood) method. The resulting tree was visualized using FigTree v1.4.4.
The detection of genetic material of the causative agents of Ebola and Marburg hemorrhagic fevers and the LCM virus was performed in serum samples using the PCR test at the Institute of Chemical Biology and Fundamental Medicine (Novosibirsk) as part of the commercial research.
The obtained results were statistically analyzed using standard methods and the GraphPad statistical analysis software. The statistical analysis of the results included calculation of the mean values, calculation of a 95% confidence interval (95% CI), evaluation of the significance of differences in mean values in the compared groups using Fisher’s exact test (differences were considered significant at 95% probability, p ≤ 0.05).
Results
Comparative analysis of optical density values in detection tests for antibodies to viruses using ELISA reagent kits with two types of conjugates
To compare the efficacy of detection of antibodies to anthroponotic viruses using ELISA with different secondary antibodies, we conducted parallel tests with conjugates to human immunoglobulins from the commercial kit and conjugates to monkey immunoglobulins. For testing, we used monkey serum panels reactive and non-reactive in the respective kits using secondary antibodies to monkey immunoglobulins. As can be seen in Table 1, the average OD450 values obtained when using conjugates to human immunoglobulins from the commercial reagent kits for detection of antibodies to herpesviruses and PI-1 and conjugates to monkey immunoglobulins did not differ significantly (≥ 0.05, the Mann–Whitney U test), thus demonstrating the interchangeability of the conjugates and the possibility of using conjugates from the reagent kits in further tests for the above markers.
Table 1. Comparative analysis of mean optical density values when detecting antibodies to herpes viruses and parainfluenza 1 virus using conjugates from commercial ELISA kits and anti-monkey secondary antibodies
Таблица 1. Сравнительный анализ средних значений оптической плотности при определении антител к герпесвирусам и вирусу парагриппа 1-го типа с использованием конъюгатов из коммерческих тест-систем и конъюгатов к иммуноглобулинам обезьян
Parameter Параметр | HSV-1,2 / ВПГ-1,2 | CMV / ЦМВ | EBV / ВЭБ | PI-1 / PI-1 | ||||
test system conjugate конъюгат тест-системы | monkey Ig conjugate конъюгат к Ig обезьян 1 : 50 000 | test system conjugate конъюгат тест-системы | monkey Ig conjugate конъюгат к Ig обезьян 1 : 50 000 | test system conjugate конъюгат тест-системы | monkey Ig conjugate конъюгат к Ig обезьян 1 : 200 000 | test system conjugate конъюгат тест-системы | monkey Ig conjugate конъюгат к Ig обезьян 1 : 2500 | |
Average OD450 values for reactive samples (n = 3) Средние значения ОП450 для реактивных образцов (n = 3) | 1.722* | 1.199* | 1.257* | 1.251* | 1.835* | 1.619* | 1.331* | 1.019* |
Average OD450 values for non-reactive samples (n = 3) Средние значения ОП450 для нереактивных образцов (n = 3) | 0.104* | 0.146* | 0.113* | 0.150* | 0.129** | 0.481** | 0.348* | 0.331* |
Note. P values were obtained by comparing the mean OD450 values between the two conjugates using the Mann–Whitney U test; * – p values ≥ 0.05 for comparative analysis; ** – p values ≤ 0.05 for comparative analysis.
Примечание. Значения р получены при сравнении средний значений ОП450 между двумя конъюгатами с использованием U-критерия Манна – Уитни; * – значения р ≥ 0,05 при сравнительном анализе; ** – значения р ≤ 0,05 при сравнительном анализе.
The samples that were non-reactive for anti-EBV were an exception: For them, the average OD450 values were significantly lower with conjugates from the reagent kits. This speaks in favor of the conjugate from the commercial DS-ELISA-ANTI-EBV-VCA-G kit compared to the RABBIT ANTI-MONKEY IgG conjugate, which produces a background in non-reactive samples even at a working dilution of 1 : 200,000.
Some of the examined animals were tested positive for markers of 6 of 11 target viruses (HSV, CMV, EBV, PI-1, enteric adenovirus, rotavirus). Table 2 summarizes detection rates for markers of anthroponotic viral infections in vervet monkeys imported from their natural habitats (Tanzania).
Table 2. Identification of markers of anthroponotic viral infections in vervet monkeys
Таблица 2. Выявление маркеров антропонозных вирусных инфекций у зеленых мартышек
№ | Virus Вирус | Marker Маркер | Number of positive samples/of examined samples Количество позитивных/исследованных образцов | % | CI 95% 95% ДИ |
Hemorrhagic fevers / Геморрагические лихорадки | |||||
1 | Ebola virus Вирус Эбола | RNA РНК | 0/75 | 0 | – |
2 | Marburg virus Вирус Марбург | RNA РНК | 0/75 | 0 | – |
3 | Lymphocytic choriomeningitis virus (LCMV) Вирус лимфоцитарного хориоменингита (ЛХМ) | RNA РНК | 0/75 | 0 | – |
Viral hepatitis / Вирусные гепатиты | |||||
4 | Hepatitis C virus (HCV) Вирус гепатита С (ВГС) | Anti-HCV* Анти-ВГС* | 0/44 | 0 | – |
Herpesvirus infections / Герпесвирусные инфекции | |||||
5 | Herpes simplex virus types 1 and 2 (HSV-1,2) Вирус простого герпеса 1-го и 2-го типов (ВПГ-1,2) | Anti-HSV-1,2 IgG Анти-ВПГ-1,2 IgG | 7/44 | 15.9 | 7–30 |
6 | Cytomegalovirus (CMV) Цитомегаловирус (ЦМВ) | Anti-CMV IgG Анти-ЦМВ IgG | 7/44 | 15.9 | 7–30 |
7 | Epstein‒Barr virus (EBV) Вирус Эпштейна‒Барр (ВЭБ) | Anti-EBV IgG Анти-ВЭБ IgG | 14/44 | 31.8 | 19–48 |
Respiratory viral infections / Респираторные вирусные инфекции | |||||
8 | Parainfluenza 1 virus (PI-1) Вирус парагриппа 1-го типа (PI-1) | Anti-PI-1 IgG Анти-PI-1 IgG | 3/44 | 6.8 | 1 – 19 |
9 | Parainfluenza 3 virus (PI-3) Вирус парагриппа 3-го типа (PI-3) | Anti-PI-3 IgG Анти-PI-3 IgG | 0/44 | 0 | – |
Intestinal viral infections / Кишечные вирусные инфекции | |||||
10 | Group A rotavirus Ротавирус группы А | Antigen Антиген | 3/21 | 14.3 | 3 – 36 |
11 | Group A–F adenovirus Аденовирусы групп A–F | DNA ДНК | 31/33 | 94 | 79 – 99 |
Note. * – a test system was used to detect total antibodies to HCV.
Примечание. * – использовалась тест-система для выявления суммарных антител к ВГС.
Hemorrhagic fevers
Since Marburg and Ebola viral hemorrhagic fevers are endemic in the African continent and pose a serious threat to humans, and LCMV, though carried asymptomatically by its natural hosts – rodents, can also cause severe viral hemorrhagic fever in monkeys and humans, the imported animals have been tested for the presence of these pathogens. The PCR test of serum samples did not detect any genetic material of these pathogens.
Virus hepatitis
Although hepatitis C is an anthroponotic infection, antibodies to structural (core) and non-structural (NS3, NS4, NS5) HCV proteins as well as IgM antibodies to the core protein were earlier detected in Old World monkeys of the genus Macaca, thus implying their possible infection with this virus or a virus antigenically similar to HCV [22]. Therefore, all the imported animals were tested for the presence of anti-HCV; however, no positive samples were detected.
Herpesvirus infections
The tests for herpesvirus infections in the green monkeys detected anti-HSV-1,2-IgG and anti-CMV-IgG at similar frequency rates – 15.9% (95% CI, 7–30%). Only one animal was tested positive for both infections. IgG antibodies to EBV were also detected in 14 (31.8%) of 44 green monkeys (95% CI, 19–48%), exceeding the detection frequency of antibodies to the two earlier mentioned viruses 2 times; however, the difference was not significant (p > 0.05). The anti-HSV-1,2 titers in positive sera ranged from 1 : 100 to 1 : 400, and the geometric mean titer was 1 : 149. The anti-CMV concentrations ranged from 0.41 to 3.63 IU/ml; the geometric mean antibody concentration was 0.91 IU/ml and the mean OD450 of antibodies to EBV was 0.953 (0.296–3.192 OD450).
Respiratory viral infections
The tests for respiratory viruses in the monkey sera detected only antibodies to PI-1 – 6.8% (95% CI, 1–19%; n = 44), while anti-PI-3 antibodies were not detected in the tested animals.
Enteric viral infections
Addressing enteric viral infections in green monkeys, we performed tests for the presence of rotavirus group A antigen and adenoviral DNA. The rotavirus antigen was detected in 14.3% (95% CI, 3–36%; n = 21) of animals, while the adenoviral DNA was detected in 94% (CI 79–99%; n = 33).
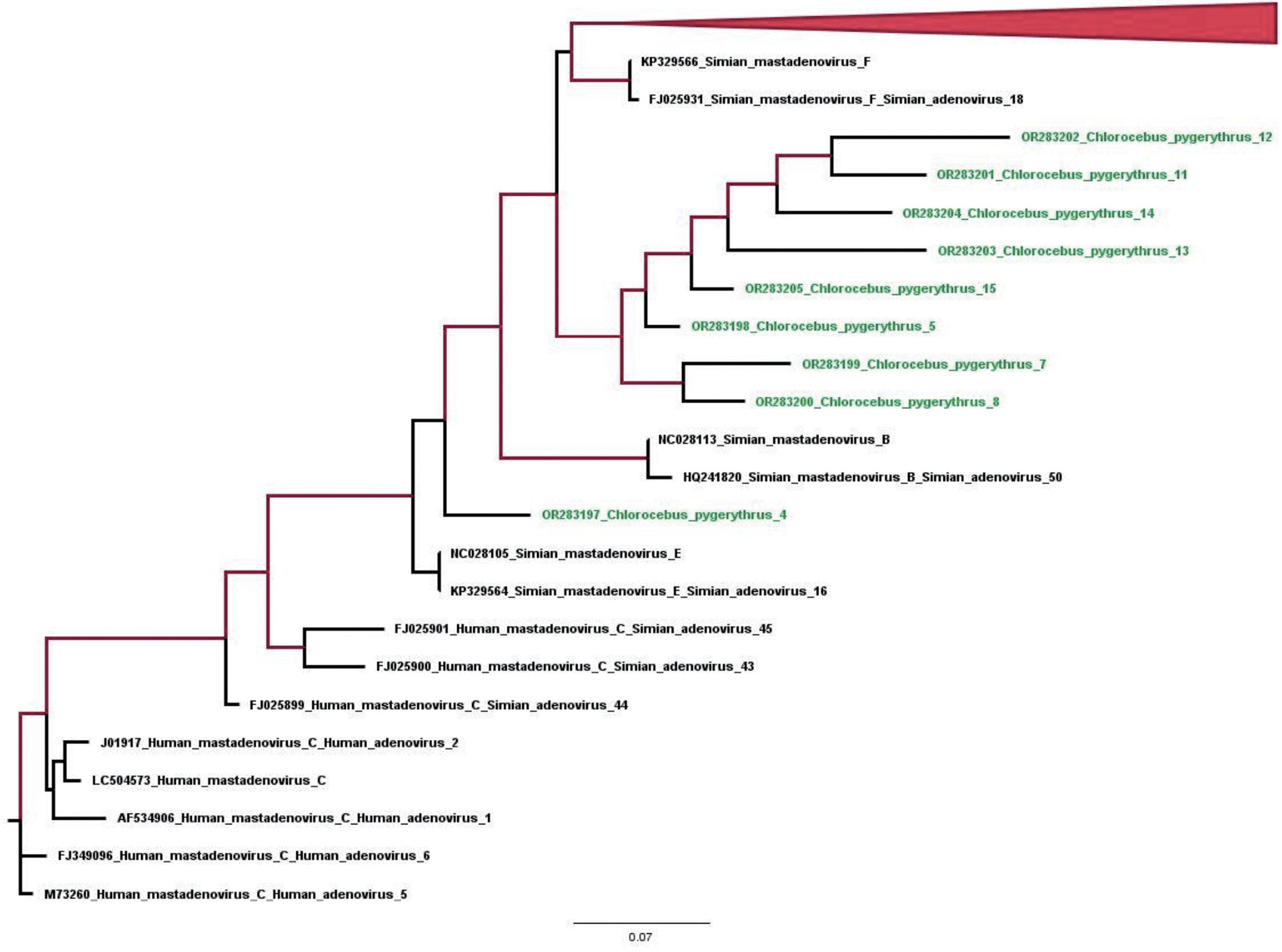
The specific detection of adenoviral DNA using primers targeting adenovirus hexon gene involved sequencing of amplified fragments 300 nucleotides long. The BLAST-based search in the NCBI database confirmed that the amplified sequences belonged to the hexon-coding region of the adenovirus genome (genome positions 17124–17424, numbering for strain KP329566 Simian mastadenovirus F). The sequences were deposited into the GenBank database (OR283197–283205); the phylogenetic analysis confirmed that the tested samples belonged to the genus Simian mastadenovirus (the family Adenoviridae), though we were not able to identify the species. In future, for the purpose of more accurate identification of the virus, we will use a set of primers that are needed to obtain the nucleotide sequence of the entire hexon gene. It should be noted that the animals tested positive for adenovirus infection did not have any clinical symptoms of enteric infection.
Discussion
The comparative analysis of mean ELISA optical density values for reactive and non-reactive samples using monkey immunoglobulin secondary antibodies and conjugates from the test systems represented by human immunoglobulin secondary antibodies demonstrated their interchangeability and the possibility to use test-system conjugates for screening of monkey sera for antibodies to herpesviruses and PI-1. Therefore, further tests were performed using conjugates from the respective test systems in accordance with the manufacturer’s protocols.
Markers for 6 of 11 studied viruses (HSV, CMV, EBV, PI-1 virus, enteric adenovirus, rotavirus) were detected among the examined animals.
Enteric infections are a major cause of morbidity and mortality in humans and animals, including monkeys. Although the gastrointestinal bacterial and parasitic pathogens and their etiological role have been thoroughly studied, there is little information about the epidemiology and spread of viral agents as well as their role in diarrheal diseases in monkeys [13].
Figure. Phylogenetic tree for the nucleotide sequences of the adenovirus genome region encoding the 281 nt hexon protein (genome posi- tions 17 122–17 403 , numbering according to strain KP329566. Simian mastadenovirus F). The tree was built using the maximum likelihood method. Branches with > 90% confidence are highlighted in red.
Рисунок. Филогенетическое дерево для нуклеотидных последовательностей участка генома аденовируса, кодирующего белок hexon величиной 281 нт (позиции генома 17 122–17 403, нумерация по штамму KP329566. Simian mastadenovirus F). Дерево построено методом максимального правдоподобия. Красным цветом выделены ветви с достоверностью более 90%.
The phylogenetic analysis of sequences of the isolated simian adenovirus of the genus Mastadenovirus apparently implies that this infection, which is not anthroponotic, circulates among animals in their natural habitats. As can be seen in the figure, all sequences differ significantly, thus providing evidence of the circulation of the above adenovirus among green monkeys in their natural habitats rather than of the infection from a single source during the quarantine or during the transportation of the captured animals. It should also be noted that the high rate of detection of asymptomatic adenovirus infection in monkeys and the confirmed zoonotic transmission require adoption of precautions in handling and maintaining primates [23, 24]. In addition, vaccine vectors derived from simian adenoviruses provide an alternative to human adenovirus vaccine vectors [25]. The detection of the rotavirus antigen in 3 animals, along with the published data of the studies [26, 27], proves that group A rotaviruses circulated among monkeys in their natural habitat, since the infection with these viruses during the quarantine or transportation would have resulted in an outbreak involving a large number of animals, considering the transmission route of the infection, as it was described previously during the HAV outbreak among these animal species [6].
Simian herpesviruses are evolutionarily closely related to human herpesviruses. Human HSV types 1 and 2 are evolutionarily related to macaque herpes B virus (Cercopithecine herpesvirus 1) and to green monkey herpesvirus 2 (Cercopithecine herpesvirus 2); human CMV – to rhesus macaque CMV (Cercopithecine herpesvirus 8); human EBV – to rhesus macaque EBV-like virus (Cercopithecine herpesvirus 15) [28]. Some of these simian viruses pose a threat to humans. Among them, special attention should be given to green monkey herpesvirus 2 also known as SA8 (simian agent 8) first isolated from green monkeys and closely related to macaque herpes B virus, the infection with which has been described in humans, including the manifestation of clinical symptoms [8].
The detection of IgG antibodies to herpesviruses in the monkeys on the 23rd day after their admission, considering the minimum contact of the animals with people while staying at the quarantine facility, suggests that the monkeys were infected with simian herpesviruses in their natural habitat. This is also confirmed by published studies describing the circulation of herpesviruses among monkeys of different species in their natural habitats [8, 28]. In addition, the average antibody titers and OD450 values in the monkey sera reactive to herpesviruses were ten times as low as the average optical density values observed in human sera reactive to these viruses [29, 30]. Considering the comparative effectiveness of detection of monkey immunoglobulins using human and monkey conjugates, such differences in optical density values can be explained by the possible antigenic cross-reactivity between monkey herpesviruses and human herpesviruses in ELISA, as it was described in published studies [8]. Thus, the detection results for antibodies to herpesviruses in monkey sera can apparently be seen as detection of antibodies to simian homologs of human herpesviruses.
Since the monkeys housed in the nursery of the Research Institute of Medical Primatology and born after 1992 do not have antibodies to the measles virus, thus being at risk of acquiring infection from imported animals, monkeys must be held in quarantine within the required period both in exporting and importing countries. Imported monkeys must also be tested for measles virus-specific IgM antibodies that indicate the recent infection. Close attention should be given to screening for markers of PI-3, which is associated with the pathology of the respiratory tract in baboons [10, 31, 32], while we have not found any published data on the role of PI-1 in the pathology of the respiratory tract in monkeys. Nevertheless, the detection of anamnestic antibodies to this virus in 3 animals most likely implies that it circulates among communities the captured monkeys were from, as the infection with this virus during the quarantine would have caused an outbreak among a larger number of animals, similar to infections caused by enteric viruses.
Conclusion
The obtained results highlight the significance of regular screening of monkeys housed in primate centers for markers of anthroponotic and zoonotic infections, including other measures aimed at prevention of any potential risk of virus circulation and cross-species transmission of viruses. The identification of new viruses among monkeys will help improve diagnostic tests of viral agents and their association with pathologies in monkeys.
Today, most primate research centers generally test imported animals upon their arrival at quarantine facilities for tuberculosis and latent viral infections to confirm their SPF status. The results of our study prove the critical importance of expanded screening for viral infections considering the epidemiological situation both in an importing country and an exporting country.
In addition, vaccination of staff members is required to confer protective immunity and to reduce the risk of transmission of socially significant infections from humans to monkeys and vice versa.
About the authors
Dmitriy I. Dogadov
Research Institute of Medical Primatology of the Ministry of Education and Science of Russia
Author for correspondence.
Email: dima_loko86@mail.ru
ORCID iD: 0000-0003-1596-0509
Ph.D. (Biol.), Researcher at the Laboratory of Infection Virology
Russian Federation, 354376, SochiKaren K. Kyuregyan
Central Research Institute of Epidemiology of the Federal Service for Surveillance on Consumer Rights Protection and Human Wellbeing; I.I. Mechnikov Research Institute of Vaccines and Sera
Email: karen-kyuregyan@yandex.ru
ORCID iD: 0000-0002-3599-117X
D.Sci. (Biol.), Professor of the RAS, Head of the Laboratory of Molecular Epidemiology, Leading Researcher at the Laboratory of Viral Hepatitis
Russian Federation, Moscow, 111123; 105064, MoscowGoncharenko M. Alexandra
Research Institute of Medical Primatology of the Ministry of Education and Science of Russia
Email: morgan_123@rambler.ru
ORCID iD: 0000-0002-6979-9784
Researcher at the Laboratory of Infection Virology, Research Institute of Medical Primatology
Russian Federation, 354376, SochiAlbert A. Minosyan
Research Institute of Medical Primatology of the Ministry of Education and Science of Russia
Email: malbert97@bk.ru
ORCID iD: 0009-0007-6459-1451
research laboratory assistant at the Laboratory of Infection Virology, Research Institute of Medical Primatology
Russian Federation, 354376, SochiArmen A. Kochkonyan
Research Institute of Medical Primatology of the Ministry of Education and Science of Russia
Email: kochkonyan7armen@gmail.com
ORCID iD: 0009-0003-1648-541X
research laboratory assistant at the Laboratory of Infection Virology, Research Institute
Russian Federation, 354376, SochiAnastasia A. Karlsen
Central Research Institute of Epidemiology of the Federal Service for Surveillance on Consumer Rights Protection and Human Wellbeing; I.I. Mechnikov Research Institute of Vaccines and Sera
Email: karlsen12@gmail.com
ORCID iD: 0000-0002-6013-7768
Researcher, Laboratory of Molecular Epidemiology of Viral Hepatitis, Researcher, Laboratory of Viral Hepatitis
Russian Federation, Moscow, 111123; 105064, MoscowOleg I. Vyshemirsky
Research Institute of Medical Primatology of the Ministry of Education and Science of Russia
Email: olegvyshem@mail.ru
ORCID iD: 0000-0002-5345-8926
Ph.D. (Biol.), Researcher at the Laboratory of Infection Virology, Research Institute
Russian Federation, 354376, SochiDzhina D. Karal-ogly
Research Institute of Medical Primatology of the Ministry of Education and Science of Russia
Email: karal_5@mail.ru
ORCID iD: 0000-0003-3606-1668
Ph.D. (Biol.), deputy director for scientific activities of the Research Institute
Russian Federation, 354376, SochiMikhail I. Mikhailov
Central Research Institute of Epidemiology of the Federal Service for Surveillance on Consumer Rights Protection and Human Wellbeing; I.I. Mechnikov Research Institute of Vaccines and Sera
Email: michmich2@yandex.ru
ORCID iD: 0000-0002-6636-6801
Ph.D. (Biol.), Professor, Academician of RAS, Chief Researcher at the Laboratory of Molecular Epidemiology of Viral Hepatitis Central Research, Head of the Laboratory of Viral Hepatitis
Russian Federation, Moscow, 111123; 105064, MoscowReferences
- Wachtman L., Mansfield K. Viral diseases of nonhuman primates. In: Nonhuman Primates in Biomedical Research. Volume 2: Diseases. Elsevier; 2012: 1–104. https://doi.org/10.1016/B978-0-12-381366-4.00001-8
- Olival K.J., Hosseini P.R., Zambrana-Torrelio C., Ross N., Bogich T.L., Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017; 546(7660): 646–50. https://doi.org/10.1038/nature22975
- Devaux C.A., Mediannikov O., Medkour H., Raoult D. Infectious disease risk across the growing human-non human primate interface: A review of the evidence. Front. Public Health. 2019; 7: 305. https://doi.org/10.3389/fpubh.2019.00305
- Patrono L.V., Samuni L., Corman V.M., Nourifar L., Röthemeier C., Wittig R.M., et al. Human coronavirus OC43 outbreak in wild chimpanzees, Côte d´Ivoire, 2016. Emerg. Microbes Infect. 2018; 7(1): 118. https://doi.org/10.1038/s41426-018-0121-2
- Liu Z.J., Qian X.K., Hong M.H., Zhang J.L., Li D.Y., Wang T.H., et al. Global view on virus infection in non-human primates and implications for public health and wildlife conservation. Zool. Res. 2021; 42(5): 626–32. https://doi.org/10.24272/j.issn.2095-8137.2021.080
- Dogadov D.I., Korzaya L.I., Karlsen A.A., Kyuregyan K.K. Molecular genetic identification of isolates of the hepatitis A virus (HAV) from monkeys at Adler Primate Center. J. Med. Primatol. 2018; 47(2): 87–92. https://doi.org/10.1111/jmp.12333
- Dogadov D.I., Korzaya L.I., Kyuregyan K.K., Karlsen A.A., Kichatova V.S., Potemkin I.A., et al. Natural infection of captive cynomolgus monkeys (Macaca fascicularis) with hepatitis E virus genotype 4. Arch. Virol. 2019; 164(10): 2515–8. https://doi.org/10.1007/s00705-019-04337-3
- Eberle R., Jones-Engel L. Understanding primate herpesviruses. J. Emerg. Dis. Virol. 2017; 3(1): 1-11. https://doi.org/10.16966/2473-1846.127
- Korzaya L.I., Dogadov D.I., Goncharenko A.M., Lapin B.A. Comparative study of anti-measles immunity in adult population of Sochi and laboratory primates of Adler primate center. Zhurnal mikrobiologii, epidemiologii i immunobiologii. 2019; 96(2): 61–7. https://doi.org/10.36233/0372-9311-2019-2-61-67 https://elibrary.ru/azxijw (in Russian)
- Korzaya L.I., Dogadov D.I., Goncharenko A.M., Karlsen A.A., Kyuregyan K.K., Mikhaylov M.I. Prevalence of laboratory markers of human respiratory viruses in monkeys of Adler primate center. Voprosy virusologii. 2022; 66(6): 425–33. https://doi.org/10.36233/0507-4088-77 https://elibrary.ru/cbntjh (in Russian)
- Molina C.V., Heinemann M.B., Kierulff C., Pissinattiet A., da Silva T.F., de Freitas D.G., et al. Leptospira spp., rotavirus, norovirus, and hepatitis E virus surveillance in a wild invasive golden-headed lion tamarin (Leontopithecus chrysomelas; Kuhl, 1820) population from an urban park in Niterói, Rio de Janeiro, Brazil. Am. J. Primatol. 2019; 81(3): e22961. https://doi.org/10.1002/ajp.22961
- Smith G.C., Lester T.L., Heberling R.L., Kalter S.S. Coronavirus-like particles in nonhuman primate feces. Arch. Virol. 1982; 72(1-2): 105–11. https://doi.org/10.1007/BF01314455
- Wang Y., Tu X., Humphrey C., McClureet H., Jiang X., Qin C., et al. Detection of viral agents in fecal specimens of monkeys with diarrhea. J. Med. Primatol. 2007; 36(2): 101–7. https://doi.org/10.1111/j.1600-0684.2006.00167.x
- Gómez J.M., Nunn C.L., Verdú M. Centrality in primate-parasite networks reveals the potential for the transmission of emerging infectious diseases to humans. Proc. Natl Acad. Sci. USA. 2013; 110(19): 7738–41. https://doi.org/10.1073/pnas.1220716110
- Wolfe N.D., Dunavan C.P., Diamond J. Origins of major human infectious diseases. Nature. 2007; 447(7142): 279–83. https://doi.org/10.1038/nature05775
- Albrecht P., Lorenz D., Klutch M.J., Vickers J.H., Ennis F.A. Fatal measles infection in marmosets pathogenesis and prophylaxis. Infect. Immun. 1980; 27(3): 969–78. https://doi.org/10.1128/iai.27.3.969-978.1980
- Choi Y.K., Simon M.A., Kim D.Y., Yoon B.I., Kwon S.W., Lee K.W., et al. Fatal measles virus infection in Japanese macaques (Macaca fuscata). Vet. Pathol. 1999; 36(6): 594–600. https://doi.org/10.1354/vp.36-6-594
- Wallis J., Lee D.R. Primate conservation: The prevention of disease transmission. Int. J. Primatol. 1999; 20(6): 803–26. https://doi.org/10.1023/A:1020879700286
- Lapin B.A., Shevtsova Z.V. Monkey viral pathology in the Sukhum colony and modeling human viral infections. J. Med. Primatol. 2018; 47(4): 273–7. https://doi.org/10.1111/jmp.12351
- Dogadov D.I., Korzaya L.I., Kyuregyan K.K., Karlsen A.A., Mikhaylov M.I. Markers of viral hepatitis E (Hepeviridae, Orthohepevirus, Orthohepevirus A) in the imported Old World monkeys. Voprosy virusologii. 2021; 66(3): 182–8. https://doi.org/10.36233/0507-4088-34 https://elibrary.ru/xvmkmz (in Russian)
- Bányai K., Esona M.D., Liu A., Wang Y., Tu X., Jiang B. Molecular detection of novel adenoviruses in fecal specimens of captive monkeys with diarrhea in China. Vet. Microbiol. 2010; 142(3-4): 416–9. https://doi.org/10.1016/j.vetmic.2009.10.014
- Korzaya L.I., Lapin B.A., Keburiya V.V., Chikobava M.G. Spontaneous infection of lower primates with hepatitis C virus. Byulleten’ eksperimental’noy biologii i meditsiny. 2002; 133(2): 178–81. https://doi.org/10.1023/A:1015511208671 https://elibrary.ru/lhjutx (in Russian)
- Kosoltanapiwat N., Tongshoob J., Ampawong S., Reamtong O., Prasittichai L., Yindee M., et al. Simian adenoviruses: Molecular and serological survey in monkeys and humans in Thailand. One Health. 2022; 15: 100434. https://doi.org/10.1016/j.onehlt.2022.100434
- Roy S., Vandenberghe L.H., Kryazhimskiy S., Grant R., Calcedo R., Yuan X., et al. Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog. 2009; 5(7): e1000503. https://doi.org/10.1371/journal.ppat.1000503
- Morris S.J., Sebastian S., Spencer A.J., Gilbert S.C. Simian adenoviruses as vaccine vectors. Future Virol. 2016; 11(9): 649–59. https://doi.org/10.2217/fvl-2016-0070
- Islam A., Hossain M.E., Haider N., Rostal M.K., Mukharjee S.K., Ferdouset J., et al. Molecular characterization of group A rotavirus from rhesus macaques (Macaca mulatta) at human–wildlife interfaces in Bangladesh. Transbound. Emerg. Dis. 2020; 67(2): 956–66. https://doi.org/10.1111/tbed.13431
- Otsyula M., Yee J., Suleman M., Tarara R., Martin J., Woods P., et al. Rotavirus infection in African, non-human primates. Ann. Trop. Med. Parasitol. 1996; 90(6): 659–61. https://doi.org/10.1080/00034983.1996.11813099
- Simmons J.H. Herpesvirus infections of laboratory macaques. J. Immunotoxicol. 2010; 7(2): 102–13. https://doi.org/10.3109/ 15476910903409843
- Goleva O.V., Murina E.A., Osipova Z.A. Serologic markers of Epstein-Barr virus reactivation in the conditions of viral encephalitis in young patients. Zhurnal infektologii. 2015; 7(1): 70–4. https://elibrary.ru/tqqpvh (in Russian)
- Makhneva N.V., Syuch N.I., Voronova V.V., Beletskaya L.V. Epstein-Barr virus and cytomegalovirus: is their role in pemphigus really incidental? A preliminary report. Al‘manakh klinicheskoy meditsiny. 2016; 44(1): 13–7. https://doi.org/10.18786/2072-0505-2016-44-1-13-17 https://elibrary.ru/vrraaj (in Russian)
- Churchill A.E. The isolation of parainfluenza 3 virus from fatal cases of pneumonia in erythrocebus patas monkeys. Br. J. Exp. Pathol. 1963; 44(5): 529–37.
- Sasaki M., Ishii A., Orba Y., Thomas Y., Hang’ombe B.M., Moonga L., et al. Human parainfluenza virus type 3 in wild nonhuman primates, Zambia. Emerg. Infect. Dis. 2013; 19(9): 1500–3. https://doi.org/10.3201/eid1909.121404
Supplementary files