A decade genetic diversity in Circulating influenza B virus in Iran (2010–2019): Divergence from WHO-recommended vaccine strains
- Authors: Emami A.1, Pirbonyeh N.2, Moattari A.2, Javanmardi F.2
-
Affiliations:
- Shiraz University of medical sciences
- Shiraz University of Medical Sciences
- Issue: Vol 68, No 5 (2023)
- Pages: 385-393
- Section: ORIGINAL RESEARCHES
- URL: https://virusjour.crie.ru/jour/article/view/12750
- DOI: https://doi.org/10.36233/0507-4088-180
- EDN: https://elibrary.ru/hqrvbk
- ID: 12750
Cite item
Full Text
Abstract
Background. Data on the disease burden and circulation patterns of influenza B virus lineages for Iran are limited.
Objective. This review aims to describe the pattern of influenza B occurrence in Iran, comparing it with the proposed vaccine strains and determining the match and mismatch with the prescribed vaccine annually.
Methods. Various sources were used to retrieve information of the data; such as information from an online search of databases such as FluNet, GISAID, and NCBI. After extracting protein sequence records in GISAID, sequence alignment with vaccine strain and construction of a phylogenetic tree were performed. Subsequently, categories of the registered circulating strains were evaluated for matching with the vaccine strains.
Results. Of the total registered influenza-positive samples, 20.21% were related to influenza B virus. The phylogenic tree was designed based on 43 samples registered in the GISAID database; 76.74 and 23.25% sequences were of Yamagata and Victoria lineages, respectively. The most prevalent influenza B virus strains circulating during the study years belonged to the Yamagata lineage. In general, the match of the influenza B virus predominant circulating strains with administrated vaccines was observed in Iran. However, a high level of mismatch between the vaccine strain and Iranian isolates was identified in 2016‒2017.
Conclusion. The review of match and mismatch in influenza vaccine in order to improve the composition of the prescribed vaccine in each region is very important because the vaccine efficacy decreased when the strain included in vaccine did not match the circulating epidemic strain.
Keywords
Full Text
Introduction
Influenza is a viral infectious disease which leads to acute complications and is considered a health concern all over the world. Among different types of influenza viruses, three species (A, B, C) are the most responsible for flu infection in humans [1]. These species are different in terms of the antigenic structure of the viral nucleoprotein (NP) and matrix protein (M), which cause various forms of severity and involve different age groups [2]. Annual epidemics are related to influenza A and B, which affect 5–10% and 20–30% of adults and children respectively each year. It is estimated that 3–5 million cases are infected with influenza type A and B which have a mortality rate of 290 000 to 650 000 each year [3, 4]. Influenza B virus is relatively more stable than influenza A virus due to its limited antigenic drift [5]. Influenza C virus is rarely reported as the cause of human diseases, likely due to the symptoms being more subclinical [6]. Although the flu subsides on its own for most people, due to the potential threat of this respiratory infection, a vaccine is the most effective method of protection recommended by health care organizations such as the World Health Organization (WHO). Due to different factors, the effect of the flu vaccine can be different. According to this, influenza (flu) vaccines cause antibodies to develop in the body which provide protection against infection with the viruses that are part of said vaccines, the protection offered by the flu vaccine varies from season to season and depends mostly on the similarity or «matching» of the viruses in the vaccine with the circulating viruses. However, it is also important to note that during the years when the flu vaccine has been matched to the circulating strain, the effect of the flu vaccine may still be different, depending on factors such as the characteristics of the person vaccinated (e.g., health, immune status and age), how the flu viruses are circulating in that season, and potentially what type of flu vaccine is used. According to the different genotypes of influenza viruses, different types of vaccine (Trivalent (H1N1, H3N2 strains, and one B strains) or quadrivalent (two B strains)) have been designed and produced [7]. Following that, a review of the genetic differentiation of circulating influenza viruses each year is required to increase the immunity of individuals who will be vaccinated, as well as design and prepare the proper vaccine type for the upcoming season. So far, many impacts of seasonal and pandemic influenza on population health have been described extensively, but due to the variable nature and diversity of influenza virus strains, it is necessary to study periodic changes and identify common strains every year in each geographical region. Although influenza A viruses cause most influenza cases and are responsible for most pandemic diseases, influenza B virus is a major cause of morbidity and mortality during interpandemic periods and its prevention is an important global health priority [8]. In the early 1980s, Influenza B virus split into two antigenically distinct phylogenetic lineages (B/Victoria/2/87, abbreviated B/Victoria-like, and B/Yamagata/16/88, abbreviated B/Yamagata-like). Based on the geographical outbreak history of type B, the Victoria lineage was limited to eastern Asia for most of the 1990s, whilst the two lineages have co-circulated the same globally from the 21st century onwards [9]. Based on the number of influenza B viruses in vaccine vials, there are two types of trivalent and quadrivalent vaccines currently available for vaccination. Due to this, the selection of influenza B virus lineage is critical to determining the effectiveness of vaccination programs. Unfortunately, the correct prediction of the predominating circulating B lineage is quite complicated and often leads to inaccuracies, resulting in a mismatch between the recommended vaccine and the circulating influenza B virus lineage. Previous studies have raised concerns that noncompliance may reduce the effectiveness of the vaccine, due to a lack of cross-protection between antigenically distinct influenza B virus lineages, leading to more influenza cases and an increase in influenza-related medical resource utilization and costs. In Iran, seasonal influenza causes many complications that place a significant economic burden on health and social systems. According to the vaccine composition recommended for the eastern Mediterranean by the WHO, during the influenza seasons in Iran, trivalent vaccines were usually used. Although high vaccination coverage levels have been reached in target vaccination groups, little is known about the effectiveness of vaccination programs in Iran. In view of the above, the most important factor about vaccine production is the surveillance data which shows virus circulation and selection based on the common influenza type during the upcoming season. The present integrative review of publicly available data aims to consolidate findings on the pattern of influenza B occurrence in Iran to have a better understanding of influenza B epidemiology and its relevance to seasonal vaccine composition.
Materials and methods
Information resources and search strategy
Various sources were used to retrieve information on epidemiological surveillance. We referred to international data sources to check WHO recommendations on the vaccine composition in Western Asia, and information on circulating influenza lineages for Iran. WHO/FluNet database was used to provide data through its network Global Influenza Surveillance and Response System (GISRS) laboratories. The data included all registered cases of influenza like illness (ILI) from sentinel sites of healthcare centers in Iran from January 2010 to December 2019, which were reported in the WHO databases weekly and FluNet web-based tool (http://www.who.int/influenza/gisrs_laboratory/flunet/en). ILI cases are defined as «a person with fever, cough and acute respiratory syndrome». During the study period, 113,319 cases of ILI were registered at FluNet databases from Iran. To determine the compatibility between the vaccine strains based on type B virus used in Iran with the circulating strains, the category of the circulating influenza viruses (Victoria or Yamagata) was appointed. َAccording to that none of the recorded samples in the FluNet database were classified after extracting the protein sequence records in GISAID, sequence alignment with vaccine strain and drawing of a phylogenetic tree was performed. After that, categories of the registered circulating strains were evaluated for matching with the vaccine strains.
Sequence data
In this cross-sectional and review study, a total of 43 HA protein sequences of influenza B viruses which were circulated during the study time in Iran with the following criteria were downloaded from the GenBank affiliated with National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/genomes/FLU/) and Global Initiative on Sharing Influenza Data (http://platform.gisaid.org/): I) Registered with Full length, II) Registered during 2010 to 2020.
The FASTA format of downloaded HA protein sequences were extracted for software analyzing.
Multiple sequence alignment and phylogenetic analysis
Reference sequences for HA protein of Influenza A virus sequences were obtained from GenBank and the EpiFlu database (Global Initiative on Sharing All Influenza Data; GISAID, https://www.gisaid.org/epiflu-applications/submitting-data-to-epiflutm/).
Multiple sequence alignments were constructed using BioEdit version 7.2.5 (http://www.mbio.ncsu.edu/bioedit/bioedit.html) with the default settings.
Phylogenetic trees were constructed by the neighbor-joining method inferred on the basis of the best fit protein substitution model for the HA protein as implemented in MEGA 7.0. The reliability of the tree was evaluated by the analysis of 1,000 bootstrap replicates and bootstrap values of > 70% were considered significant. Designated clades were chosen based on the clustering patterns in the HA phylogeny.
Analyses
Epidemiological profiles of the positive cases (week of occurrence, year, and type of infection) are presented during the time of the study. Data of Iranian isolates, reference strain, and vaccine strain were collected and consolidated based on the influenza database. For the alignment of protein sequences and strain match-mismatch with vaccine strains MEGA7 software was used (https://www.megasoftware.net/).
Ethics
The study procedures were reviewed by relevant ethical committees affiliated with Shiraz University of Medical Sciences and approved by following Ethical code: IR.SUMS.REC.1398.1173.
Results
Strain data of Influenza viruses
Epidemiological monitoring systems
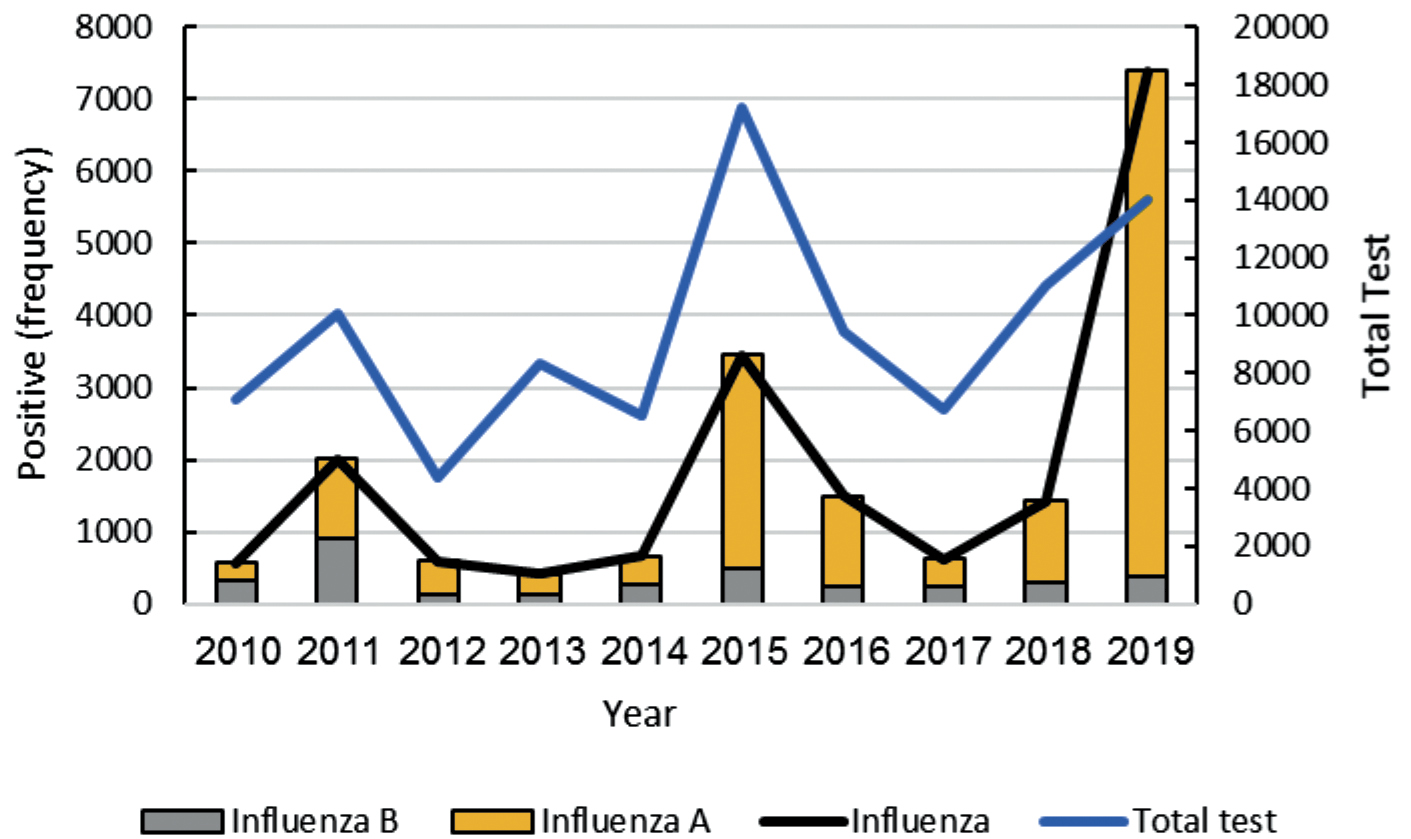
A total of 113,319 samples pattern of Iranian as ILI cases were extracted from the WHO/FluNet database on influenza A and B virus subtypes (Epidemiological, 2010 to 2019). Among these, 19,333 samples were reported influenza-positive which 3,908 (3,908/19,333, 20.21%) samples were categorized as influenza B virus (lineage is not determined). Based on the assessments it is determined that the flu (A and B) outbreaks were mainly during 2011, 2015 and 2019 years (Fig. 1, Blackline). This is while in the year 2011, influenza B was more frequent.
Phylogenic tree
Due to being unable to determine the subtype of influenza B virus in the FluNet database, in order to identify the lineage of circulating the influenza B virus in Iran during study years, the phylogenic tree was built based on 43 samples registered in GISAID database. According to this and based on the HA protein sequence of samples, subtypes of influenza type B lineage were determined. Among these samples, 33 (76.74%) were identified as influenza B virus strain/lineage Yamagata and 10 (23.25%) – as influenza B virus strain/lineage Victoria (Table 1).
Table 1. Frequency of influenza B virus lineage
Years of Study | |||||||||||
2010 | 2011a | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | 2019 | Total | |
B/Victoria lineage | 2 (50%) | – | 2 (40%) | – | – | 1 (20%) | 2 (100%) | 1 (9.09%) | – | 2 (40%) | 10 |
B/Yamagata lineage | 2 (50%) | – | 3 (60%) | 4 (100%) | 5 (100%) | 4 (80%) | – | 10 (90.9%) | 2 (100%) | 3 (60%) | 33 |
Total influenza B | 4 | – | 5 | 4 | 5 | 5 | 2 | 11 | 2 | 5 | 43 |
Note. aAccording to reports, influenza B virus was rarely circulating in Iran in 2011, but not reported in the GISAID database.
Fig. 1. Annual flu (A and B) circulation extracted from the WHO/FluNet database in the last decade.
Based on the phylogenic tree, it was found that the most prevalent circulating influenza B viruses during study years belonged to the Yamagata lineage (Fig. 2). In following assessments, although Influenza type B/Victoria lineage had been more predominant in the year 2016, it was circulating with the Yamagata lineage for most of the years. Following the phylogenetic analysis of the HA protein sequences, one and two genetic clades of influenza B/Victoria lineage and influenza B/Yamagata lineage were identified respectively. According to reports, influenza B virus was rarely circulating in Iran in 2011, but not reported in the GISAID database (Table 1).
Fig. 2. Cladogram of influenza B virus isolated during the study time.
In order to check the genetic changes of circulating influenza B virus strains, B/Brisbane/60/2008, B/Massachusetts/02/2012, and B/Phuket/3073/2013 strains as the eastern Mediterranean recommended reference strains by WHO, were used.
Victoria Lineage specifications
Based on the phylogenic tree, all the detected Victoria strains in Iran were categorized in clade 1A, except the years 2011, 2013,2014, and 2018 when no Victoria lineage was detected. In comparing circulating Victoria strains with B/Brisbane/60/2008 vaccine strains, a series of amino acid changes were observed in the HA1 region. In all Victoria isolate sequences collected in the years 2010 and 2012 amino acid substitution I146V have been detected, whereas in the 2015–2017 and 2019 isolates amino acid substitutions I117V and N129D have been identified. The additional amino acid substitutions, V87A and I175V, have been observed in 2015 and 2017 isolates. In isolates of the year 2019, 3 amino acid deletions have been seen in positions D164, K165, and N166, while additional substitution N129D have been seen in sequence as well.
Yamagata Lineage specifications
Based on the assessment of the phylogenic tree for influenza B/Yamagata lineage, all isolates were categorized into clades 2 and 3. According to the analysis of results, the most circulating isolates of influenza B/Yamagata lineage belonged to clade 3. This is while two of the most isolates in the year 2012; B/Ghom/28441/2012, and B/Yazd/9800/2012, have been categorized in clade 2. To compare circulating Yamagata strains and assess the changes, two vaccine strains, B/Massachusetts/02/2012 (2013) for clade 2 and B/Phuket/3073/2013 (2015 and 2019) for clade 3 were used. These two strains have several differences in their HA nucleotide sequence as following: K48R, A108P, N116K, S150I, N166Y, A182T, N203S, G230D, K299E, and E313K. In comparing Yamagata circulating isolates sequences in clade 2 with reference strain, isolates of the year 2012 showed amino acid substitutions in D196N, T121S, and G141R regions.
All Iranian isolates from 2013 onwards were mostly placed in the Yamagata lineage. b/Ghom/2844/2012 and b/Yazd/9800/2012 strains were classified in clade 2. The exclusive mutation in these strains was D196N, in addition, amino acid substitutions G141R and T121S were observed in Ghom strain and Yazd strain, respectively. Mutations of isolates compared to vaccine strains are shown on the phylogenetic tree. The most prevalent mutations in Iranian strains of Yamagata lineage were X198T, L172Q, and M251V.
The phylogenetic tree was generated for HA protein sequence by the neighbor-joining method with 1,000 bootstrap replicates. Branch values > 70 are indicated. The scale bar represents approximately 1% protein change between close relatives.
Results of matching analysis with vaccine strains
Table 2. Comparison of influenza B virus vaccine and circulating lineage, Iran, 2010–2019 (WHO / FluNet)
Reference | Predominant lineage (n; %)a | WHO Recommended Vaccine | % Mismatchb |
2010 | Victoria = Yamagata (2; 50) | B/Brisbane/60/2008 (B/Victoria lineage) | 50% |
2011c | – | B/Brisbane/60/2008 (B/Victoria lineage) | − |
2012 | Yamagata (3; 60) | B/Wisconsin/1/2010 (B/Victoria lineage) | 60% |
2013 | Yamagata (4; 100) | B/Massachusetts/2/2013 (B/Yamagata lineage) | 0 |
2014 | Yamagata (5; 100) | B/Massachusetts/2/2013 (B/Yamagata lineage) | 0 |
2015 | Yamagata (4; 80) | B/Phuket/3073/2013-like virus (B/Yamagata/16/88 lineage) | 20% |
2016 | Victoria (2; 100) | B/Phuket/3073/2013-like virus (B/Yamagata/16/88 lineage) | 100% |
2017 | Yamagata (10; 90.9) | B/Brisbane/60/2008 (B/Victoria lineage) | 91.9% |
2018 | Yamagata (2; 100) | B/Phuket/3073/2013-like virus (B/Yamagata/16/88 lineage) B/Colorado/06/2017-like virus (B/Victoria/2/87 lineage) | − |
2019 | Yamagata (3; 60) | B/Phuket/3073/2013-like virus (B/Yamagata/16/88 lineage) B/Colorado/06/2017-like virus (B/Victoria/2/87 lineage) | − |
Note. aThe percentage are representative of the dominant lineage which are estimated by the total number of samples positive for influenza B virus as the denominator; b% Mismatch = 100% − % Circulating B lineage match with vaccine; cinfluenza B virus was rarely circulating in Iran in 2011, and not reported in the GISAID database.
Table 2 shows the matching of the predominant circulating strains of influenza B virus with administrated vaccines during the study time in Iran (2010–2019) based on WHO/FluNet reports.
Based on this analysis, high levels of mismatch (100 and 91.9%) are observed in the years 2016 and 2017 respectively.
Discussion
In the present work, we conducted a 10 year review to estimate the protection coverage of administered influenza vaccines in Iran. The current results showed a lot of mismatches (≥ 50%) between the years 2010−2017 in administrated vaccines and circulating influenza virus lineages.
As we know the genome of influenza A and B viruses are segmented and each contains 8 gene segments. Influenza A viruses based on the antigenic properties of their surface glycoproteins are further divided into 16 hemagglutinin (H1–16) and 9 neuraminidase (N1–9) subtypes [10]. Two influenza A subtypes (H1N1 and H3N2) and two antigenically distinct lineages of influenza B viruses (Yamagata and Victoria) are currently co-circulating in humans [3]. Based on the WHO recommendation, vaccination is the best method for the prevention and control of influenza. Vaccination can diminish illness and reduce the severity of influenza infection, particularly in at-risk groups including young children and the elderly [11]. To be focused on the effectiveness of licensed influenza vaccines, current seasonal vaccines required annual evaluation and reformulation to keep the rate with the antigenic drift of circulating strains. To select the vaccine composition for the upcoming season, a routine evaluation is carried out twice a year; once, usually in February, to select strains for the Northern hemisphere vaccine, and again, typically in September, for the Southern hemisphere [12]. To accommodate the steps of vaccine production, this process is performed 7 to 8 months in advance of the «flu season» [3]. Although the selection of vaccine strains to include in annual influenza vaccines is based on global surveillance of circulating influenza viruses, but according to various evidence there are a lot mismatches in several countries [13−15]. This may be due to the following reasons: I.) Impossibility to study the annual circulating strains, II.) Negligence to perform diagnostic tests in a timely manner, III.) Lack of attention and trust of WHO influenza collaboration centers in test results, IV.) Lack of an accurate registry system and correct information on the success rate of vaccination. The seasonal patterns described in the current study in Iran further support a proposed influenza vaccination schedule during October and November. Assessment of the recorded global results shows that a change in the relative predominance of the two influenza B lineages is essentially observed every two/three years [16, 17]. In some systematic reviews it has been shown that both influenza B virus lineages are co-circulating in different years [8]. Lack of knowledge about the circulating influenza strains in some countries makes it impossible for health organizations to prepare and purchase the appropriate vaccine for injection during the flu season outbreak. This also means that despite the high vaccination coverage rate, vaccine does not provide adequate safety coverage in the target population. One of the important threats in vaccine incompatibility with the circulating strains in a country is often a widespread epidemic. In addition to the causes mentioned in the occurrence of mismatch in the administered vaccine is the emergence of viruses with novel antigenic properties. An example of this was the 2009 pandemic (A(H1N1)pdm09), which the difficulty in producing and distributing a proper vaccine in a short timeframe caused the second wave of the pandemic [18, 19]. In this review, we aimed to summarize the available evidence based on influenza B disease patterns in Iran as well as the evidence in the global health ministry databases. Following that, we tried to find the mismatches between vaccine types and circulating strains of the influenza B virus in the timespan of a decade. Due to the analysis of our study results, it has been shown that both influenza B virus lineages (B/Yamagata and B/Victoria) have been co-circulating in most years in the influenza season. Similar to the results of our study, phylogenetic analysis of influenza B viruses circulating in Italy over the entire study period confirmed the co-circulation of both lineages during each season and revealed a mixed circulation of distinct evolutionary viral variants with different matching levels to the vaccine strains. In particular, a gradual drift was observed in both lineages and further clades along with subclades were identified in each lineage [20]. Meanwhile, in the year 2011 no influenza B was reported, and in the year 2016, only Victoria strain had been reported for 3 consecutive years. Based on analysis of limited recorded information, although a lineage switch occurred each year, the B/Yamagata strain was the most prevalent circulating strain in Iran, according to the reports that have shown it being the only common strain for many years. Furthermore, this date indicates a high degree of mismatch between the vaccine and the predominant circulating strain (91.9–100%) in 2016–2017, and a partial mismatches during the other influenza seasons.
Based on different experiences, selection of the influenza type B virus lineage is considered critical in determining the effectiveness of vaccination programs [21]. Unfortunately, improper prediction of the predominant circulating B lineage, often leads to inaccuracies and causes a mismatch between the administered vaccine and the circulating influenza B virus strain. In previous experiences, it was a concern that mismatches occur due to the absence of cross-protection between antigenically distinct influenza B viruses, which can result in lower vaccine effectiveness, leading to more influenza cases [22, 23]. This burden of the influenza infection will increase the influenza-related medical resources utilization and costs. Due to the importance of this point, it has been found that the degree of vaccine compliance with the circulating influenza B virus during the flu season is effective in vaccination programs [24]. Although high vaccination coverage levels have been reached in target groups in Iran, little is known about the effectiveness of vaccination programs, and data on the burden of influenza disease are limited as well, which is most likely due to a lack of reporting. In the current study, based on WHO/FluNet registered data, moderate levels of influenza B burden (13.77–54.67%), except for 2019 (4.99%) were observed over the first nine years of study time. This displacement of influenza B circulation could potentially be because of the COVID-19 (SARS-CoV-2) pandemic in the year 2019. In 2019, the use of personal protective equipment such as masks and the presence of strict quarantine conditions in almost all countries greatly reduced the prevalence of respiratory viral infection such as influenza, especially the influenza B virus [25]. According to the results of the present study, in most years of research (70%), there is a mismatch between the influenza B virus circulating strains and the administered vaccine strains. Due to the time of influenza B outbreak in Iran (From December to the end of March) and the existence of numerous research capacities in universities and research centers of the country (based on the numerous published articles), unfortunately, limited results of annually circulated strains (present only in a few large cities) have been recorded in international database systems such as WHO/FluNet from Iran. Given the vastness of Iran and hostile sanctions, this has led to different parts of the country not having access to the necessary facilities and equipment for sampling and inspection of circulating isolates in due time. Also, the limitation of diagnostic facilities, even in the major centers of the country, has in turn, limited the possibility to perform multiple tests and repeat the confirmation. Failure to register the circulating virus information in a timely manner will result in a lack of timely action in preparing appropriate vaccinations and a lack of complete safety coverage in the country. In addition to creating an epidemic in the country, this could also be a threat of the disease spreading through the whole region. Given that the unpredictability of influenza B lineage circulation during the years causes the mismatch in influenza season with vaccine strain, the disease burden (in terms of clinical cases, hospitalizations, and health care resource utilization) and the societal burden can be considerable. Also, it is estimated to have cost health care providers and society well over several billion. In addition, international threats of an outbreak could add to the scale of the problem. Considering the above-mentioned cases, and the problem of non-compliance in many other countries including Taiwan and Brazil [26, 27] it is worth mentioning that in addition to the problems raised, some cases can be implemented and reduced with the planning and support of related organizations.
Conclusion
Considering the non-prescription of quadrivalent vaccines due to the lack of access and double the financial burden on the health system before 2018 in Iran and the need to justify the change of consumed vaccines and the prescription of quadrivalent vaccines in high-risk groups such as people with weak immune systems, health care workers, pregnant women, the elderly, and children multiply the need for quadrivalent vaccines due to the incompatibility of the vaccines used.
Also, influenza B viruses can circulate during a pandemic, allowing for the re-emergence of old lineages due to reassortment between different strains. As a result, there is a need to improve laboratory-based influenza surveillance in the world and help improve other developing countries such as Iran as the largest country in the Middle East region. Another important point in influenza vaccination is that the existing influenza B vaccine is limited in that it does not provide cross-protection between two distinct strains of influenza B virus, and as a result, vaccine efficacy is reduced when the introduced vaccine strain is mismatched with the circulating epidemic strain. This calls for an intensive, extensive and focused study of the changes in the circulating virus globally.
Recommendation
There were some restrictions such as limited confirmed data in our study. Only protein sequences registered in GISAID, were included in the study and analyzed by the phylogenic tree as the confirmed lineage. This could underestimate the real proportion of circulating viruses and the true incidence of influenza infections in the population. Nevertheless, we can assume that these results would largely reflect the circulation of the two main influenza B virus lineages in Iran.
About the authors
Amir Emami
Shiraz University of medical sciences
Author for correspondence.
Email: emami.microbia@gmail.com
ORCID iD: 0000-0002-4510-1820
Ph.D (Microbiology), Department of Microbiology, Burn & Wound Healing Research Center
Iran, Islamic Republic of, ShirazNeda Pirbonyeh
Shiraz University of Medical Sciences
Email: pirbonyeh@yahoo.com
ORCID iD: 0000-0001-5700-3913
Department of Microbiology, Burn & Wound Healing Research Center, Department of Bacteriology and Virology
Iran, Islamic Republic of, ShirazAfagh Moattari
Shiraz University of Medical Sciences
Email: moattari.a@sums.ac.ir
ORCID iD: 0000-0003-4394-9622
Ph.D. (Bacteriology and Virology), Professor of Medical Virology, Department of Bacteriology and Virology
Iran, Islamic Republic of, ShirazFatemeh Javanmardi
Shiraz University of Medical Sciences
Email: javanmardi.biostat@yahoo.com
ORCID iD: 0000-0001-8841-0861
кандидат наук (биостатистика), кафедра биостатистики
Iran, Islamic Republic of, ShirazReferences
- Chen H., Park S.G., Choi N., Moon J.I., Dang H., Das A., et al. SERS imaging-based aptasensor for ultrasensitive and reproducible detection of influenza virus A. Biosens. Bioelectron. 2020; 167: 112496. https://doi.org/10.1016/j.bios.2020.112496
- Tavakoli F., Moattari A., Shamsi Shahr Abadi M., Kadivar M.R., Khodadad N., Pirbonyeh N., et al. Antigenic variation of the haemagglutinin gene of the influenza A (H1N1) pdm09 virus circulating in Shiraz, February-April 2013. Iran. J. Immunol. 2015; 12(3): 198–208.
- Houser K., Subbarao K. Influenza vaccines: challenges and solutions. Cell Host Microbe. 2015; 17(3): 295–300. https://doi.org/10.1016/j.chom.2015.02.012
- WHO. Influenza (Seasonal). Available at: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal)
- Kim H., Webster R.G., Webby R.J. Influenza virus: dealing with a drifting and shifting pathogen. Viral Immunol. 2018; 31(2):174–83. https://doi.org/10.1089/vim.2017.0141
- CDC. 2021-2022 U.S. Flu Season: Preliminary In-Season Burden Estimates. Available at: https://www.cdc.gov/flu/about/burden/2021-2022.htm#2021-burden-est
- Gaglani M., Vasudevan A., Raiyani C., Murthy K., Chen W., Reis M., et al. Effectiveness of trivalent and quadrivalent inactivated vaccines against influenza B in the United States, 2011-2012 to 2016-2017. Clin. Infect. Dis. 2021; 72(7): 1147–57. https://doi.org/10.1093/cid/ciaa102
- Paul Glezen W., Schmier J.K., Kuehn C.M., Ryan K.J., Oxford J. The burden of influenza B: a structured literature review. Am. J. Public Health. 2013; 103(3): e43-51. https://doi.org/10.2105/ajph.2012.301137
- Rota P.A., Wallis T.R., Harmon M.W., Rota J.S., Kendal A.P, Nerome K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990; 175(1): 59–68. https://doi.org/10.1016/0042-6822(90)90186-u
- McAuley J.L., Gilbertson B.P., Trifkovic S., Brown L.E., McKimm-Breschkin J.L. Influenza virus neuraminidase structure and functions. Front. Microbiol. 2019; 10: 39. https://doi.org/10.3389/fmicb.2019.00039
- Flannery B., Clippard J., Zimmerman R.K., Nowalk M.P., Jackson M.L., Jackson L.A., et al. Early estimates of seasonal influenza vaccine effectiveness – United States, January 2015. MMWR Morb. Mortal. Wkly Rep. 2015; 64(1): 10–5.
- Monto A.S., Petrie J.G. Improving influenza vaccine effectiveness: ways to begin solving the problem. Clin. Infect. Dis. 2019; 69(10): 1824–6. https://doi.org/10.1093/cid/ciz416
- Tricco A.C., Chit A., Soobiah C., Hallett D., Meier G., Chen M.H., et al. Comparing influenza vaccine efficacy against mismatched and matched strains: a systematic review and meta-analysis. BMC Med. 2013; 11: 153. https://doi.org/10.1186/1741-7015-11-153
- Flannery B., Kondor R.J.G., Chung J.R., Gaglani M., Reis M., Zimmerman R.K., et al. Spread of antigenically drifted influenza A (H3N2) viruses and vaccine effectiveness in the United States during the 2018–2019 season. J. Infect. Dis. 2020; 221(1): 8–15. https://doi.org/10.1093/infdis/jiz543
- Paules C.I., Fauci A.S. Influenza vaccines: good, but we can do better. J. Infect. Dis. 2019; 219(Suppl. 1): S1–4. https://doi.org/10.1093/infdis/jiy633
- Update: influenza activity – United States, 2010-11 season, and composition of the 2011-12 influenza vaccine. MMWR Morb. Mortal. Wkly Rep. 2011; 60(21): 705–12.
- Jackson D., Elderfield R.A., Barclay W.S. Molecular studies of influenza B virus in the reverse genetics era. J. Gen. Virol. 2011; 92(Pt. 1): 1–17. https://doi.org/10.1099/vir.0.026187-0
- Lee Y., Kim K., Ko E., Lee Y., Kim M., Kwon Y., et al. New vaccines against influenza virus. Clin. Exp. Vaccine Res. 2014; 3(1): 12–28. https://doi.org/10.7774/cevr.2014.3.1.12
- Use of influenza A (H1N1) 2009 monovalent vaccine: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2009. MMWR Recomm. Rep. 2009; 58(RR-10): 1–8.
- Puzelli S., Di Martino A., Facchini M., Fabiani C., Calzoletti L., Di Mario G., et al. Co-circulation of the two influenza B lineages during 13 consecutive influenza surveillance seasons in Italy, 2004–2017. BMC Infect. Dis. 2019; 19(1): 990. https://doi.org/10.1186/s12879-019-4621-z
- WHO Expert Committee on Biological Standardization: Fifty-fourth Report; 2005.
- Skowronski D., Masaro C., Kwindt T., Mak A., Petric M., Li Y., et al. Estimating vaccine effectiveness against laboratory-confirmed influenza using a sentinel physician network: results from the 2005-2006 season of dual A and B vaccine mismatch in Canada. Vaccine. 2007; 25(15): 2842–51. https://doi.org/10.1016/j.vaccine.2006.10.002
- Carrat F., Flahault A. Influenza vaccine: the challenge of antigenic drift. Vaccine. 2007; 25(39-40): 6852–62. https://doi.org/10.1016/j.vaccine.2007.07.027
- Luna E.J.A., Gattás V.L., Campos S.R.S.L.C. Effectiveness of the Brazilian influenza vaccination policy: a systematic review. Epidemiol. Serv. Saúde. 2014; 23(3): 559–75. https://doi.org/10.5123/S1679-49742014000300020
- Olsen S.J., Azziz-Baumgartner E., Budd A.P., Brammer L., Sullivan S., Pineda R.F., et al. Decreased influenza activity during the COVID-19 pandemic – United States, Australia, Chile, and South Africa, 2020. Am. J. Transplant. 2020; 20(12): 3681–5. https://doi.org/10.1111/ajt.16381
- Barros E.N., Cintra O., Rossetto E., Freitas L., Colindres R. Patterns of influenza B circulation in Brazil and its relevance to seasonal vaccine composition. Braz. J. Infect. Dis. 2016; 20(1): 81–90. https://doi.org/10.1016/j.bjid.2015.09.009
- Okoli G.N., Racovitan F., Righolt C.H., Mahmud S.M. Variations in seasonal influenza vaccine effectiveness due to study characteristics: a systematic review and meta-analysis of test-negative design studies. Open Forum Infect. Dis. 2020; 7(7): ofaa177. https://doi.org/10.1093/ofid/ofaa177
Supplementary files