Unusual BA222-like strains of Rotavirus A (Sedoreoviridae: Rotavirus: Rotavirus A): molecular and genetic analysis based on all genome segments
- Authors: Velikzhanina E.I.1, Sashina T.A.1, Morozova O.V.1, Kashnikov A.Y.1, Epifanova N.V.1, Novikova N.A.1
-
Affiliations:
- Academician I.N. Blokhina Nizhny Novgorod Scientific Research Institute of Epidemiology and Microbiology of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
- Issue: Vol 69, No 4 (2024)
- Pages: 363-376
- Section: ORIGINAL RESEARCH
- URL: https://virusjour.crie.ru/jour/article/view/16664
- DOI: https://doi.org/10.36233/0507-4088-254
- EDN: https://elibrary.ru/ogoquq
- ID: 16664
Cite item
Abstract
Introduction. Rotavirus infection is the major cause of severe dehydrating diarrhea requiring hospitalization in young children worldwide. Due to their segmented genome, rotaviruses are capable of gene reassortment, which makes the emergence and spread of genetically novel strains possible. The purpose of this study was to search for unusual rotaviruses circulating in Nizhny Novgorod in 2021‒2023 and their molecular genetic characterization based on all genome segments.
Materials and methods. Rotavirus-positive stool samples of children were examined by PCR genotyping and electrophoresis in PAAG. cDNA fragments of each of the 11 genes (VP1‒VP4, VP6, VP7, NSP1‒NSP5), 570 to 850 nucleotide pairs in length were sequenced for the selected strains. The phylogenetic analysis was performed in the MEGA X program.
Results. In the study period 2021‒2023, 11 G[P] combinations with a predominance of G3P[8] (59.5%) were identified. Six atypical Rotavirus А (RVA) strains were identified: 2 strains of the G2P[4] genotype (G2-P[4]-I2-R2-C2-M2-A3-N2-T3-E2-H3, G2-P[4]-I2-R2-C2-M2-A3-N2-T3-E3-H2) and 4 G3P[9] strains (all strains had the genotype G3-P[9]-I2-R2-C2-M2-A3-N2-T3-E3-H3). Phylogenetic analysis based on all genes showed an evolutionary relationship between rotaviruses similar to rotaviruses of cats and dogs (BA222-like) and unusual strains of the G2P[4] genotype, for which a mixed combination of genotypes was identified and characterized for the first time.
Discussion. The results obtained expand the understanding of the diversity of reassortant RVAs, as well as complement the data on the genotypic structure of the rotavirus population in Nizhny Novgorod.
Conclusion. The wide genetic diversity of reassortant RVA can help rotaviruses overcome the immunological pressure provided by natural and vaccine-induced immunity. In this regard, to control the emergence of new variants and assess changes in the virulence of rotaviruses after reassortment processes, continuous molecular monitoring for circulating RVA is necessary.
Full Text
Introduction
Currently, most intestinal infections of established etiology are caused by viral pathogens. The spectrum of intestinal viruses is very diverse, but the leading cause of incidence among children is Rotavirus A (RVA, Rotavirus genus, Sedoreoviridae family, Reovirales order, Resentoviricetes class, Duplornaviricota type). Rotavirus infection (RVI) accounts for approximately 228,000 deaths worldwide each year, including approximately 128,500 among children under 5 years of age [1, 2]. The greatest burden of RVI is among children living in low- and middle-income countries, especially because of unfavorable living conditions, limited supply of clean drinking water and poor sanitation [3]. In Russia, despite the annually increasing number of children vaccinated against RVI, the vaccination coverage of the target cohort remains extremely low to influence the epidemic process (2020 – 3.68%; 2021 – 6.23%; 2022 – 7.15%; 2023 – 12.7%) [4, 5].
A distinctive feature of rotaviruses (RVs) is a segmented genome consisting of 11 segments of double-stranded RNA with a total length of about 18,555 nucleotides encoding 6 structural (VP1–VP4, VP6–VP7) and 6 non-structural (NSP1–NSP6) proteins [6–8]. RVs are characterized by a wide antigenic and genetic diversity. The VP4 and VP7 genes have been most fully characterized, on the basis of which G- and P-genotypes are determined using binary classification [9]. To date, 42 G-genotypes and 58 P-genotypes are known for RVA [10]. The most common in the world, including Russia, are considered to be 6 combinations of G[P]-types of RVA: G1P[8], G2P[4], G3P[8], G4P[8], G9P[8], and G12P[8], which account for approximately 90% of cases of RVA infection [11, 12]. The number of less common G[P]-variants is much larger, but they account for only 4.9% of infections. For example, they include such genotypes as G9P[4], G9P[9], G8P[8], G2P[8], G4P[4], G3P[9], and others [13‒16].
The binary classification system focuses only on the VP4 and VP7 gene segments and does not provide information on the diversity of other genes. Therefore, to better understand the epidemiology and evolution of RV, a full-genome classification system was proposed to assign each segment of the virus genome to a specific genotype. To describe the complete genotype, the label Gx-P[x]-Ix-Rx-Rx-Cx-Mx-Ax-Nx-Tx-Ex-Hx is used for genes encoding VP7-VP4-VP6-VP1-VP2-VP3-NSP1-NSP2-NSP3-NSP4-NSP5/NSP6 proteins, respectively [17]. Determination of the complete genotype facilitates the detection of rare and unusual RVA and provides a better understanding of the origin and diversity of strains, which, in turn, is important for successful prediction of the epidemic situation and assessment of the impact of vaccine prophylaxis on the pathogen population.
Based on whole-genome typing, 3 genetic groups are distinguished among RVAs. Worldwide, the frequently occurring combinations of G1, G3, G4, G9, G12, and P[8] are usually associated with the Wa-like (1st) genetic group (G1/G3/G4/G9-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1), which is distantly related to porcine RVs [18, 19]. Such RVs are characterized by a long migration profile of genome segments in polyacrylamide gel electrophoresis (PAGE). G2P[4] genotype strains, in turn, belong to the DS-1-like (2nd) genetic group (G2-P[4]-I2-R2-C2-M2-A2-N2-T2-E2-H2), evolutionarily related to bovine RVs [17, 19]. They have a short genomic segment migration profile. There is a 3rd, minor, AU-1-like genetic group (G3P[9]-I3-R3-R3-C3-M3-A3-A3-N3-T3-E3-H3), which includes strains similar to cat and dog RVs characterized by a broad segment migration profile [20, 21]. Strains with a pure AU-1-like combination are rarely found in humans, while variants with a mixed set of AU-1-like and DS-1-like genes are more common. They are usually assigned to a separate group, the prototype for which is RVA strain VA222 (G3-P[9]-I2-R2-R2-C2-C2-M2-A2-A3-N1-T3-E2-H3) isolated from a domestic cat in Italy in 2005 [22, 23].
The presence of a segmented genome determines the ability of RV to exchange segments (reassortment) when two or more strains simultaneously infect a cell, which allows genes with different specificity to group independently of each other, thus maintaining diversity in the population. The spread of epidemically significant reassortant strains of RVA, which carry genetic material of several genetic groups, has been increasingly observed worldwide [9, 24, 25].
In this regard, the aim of this study was to search for unusual RVs in Nizhny Novgorod and their molecular genetic characterization based on all genome segments.
Materials and methods
Stool samples from children hospitalized in a children’s infectious disease hospital in Nizhny Novgorod with symptoms of acute intestinal infection in the period 2021–2023 were used.
Nucleic acid extraction and polymerase chain reaction (PCR) with reverse transcription (RT-PCR) were performed using RIBO-prep and REVERTA-L reagent kits (Central Research Institute of Epidemiology of Rospotrebnadzor (CRIE), Russia). Rotavirus RNA was detected using AmpliSense Rotavirus/Norovirus/Astrovirus FL and AmpliSense Viro-Screen-FL real-time PCR test systems (CRIE). Additionally, viral RNA was analyzed by polyacrylamide gel electrophoresis (RNA-PAAG) [26].
G[P]-genotype of RV was determined by multiplex PCR based on primers specific for genotypes G1–G4, G6, G8, G9, G12, P[4], P[6], P[8], and P[9] [26–32]. Results were detected by electrophoresis in agarose gel containing ethidium bromide.
For each gene, one complementary DNA (cDNA) fragment of 570 to 850 nucleotide pairs in length was obtained using primers published previously [32]. The cDNA fragments were sequenced along two strands using forward and reverse primers on a Nanofor 05 instrument (Institute of Analytical Instrumentation (IAP), Russian Academy of Sciences, Russia) using the BigDye Terminator v3.1 sequencing kit (Thermo Fisher Scientific, USA). Nucleotide sequences are available in the GenBank database under the numbers PP475712–PP475777.
The search for related sequences was performed using the BLAST online service. For phylogenetic analysis, nucleotide sequences of 11 RV genes circulating in different countries, including Russia (Novosibirsk, Omsk and Moscow), as well as RV sequences from Nizhny Novgorod obtained earlier, were retrieved from GenBank. The GenBank registration numbers of these strains are given in the names of the isolates on the phylogenetic trees. Nucleotide sequence alignment and analysis were performed in the MEGA X program version 10.0.5. Phylogenetic trees were constructed using the Maximum Likelihood method [33, 34]. Bootstrap analysis was performed based on 1000 random samples. The percentages of nucleotide sequence similarity were calculated using the Pairwise Distances method. The affiliation of the studied strains to phylogenetic lineages and sublineages was determined based on clustering of isolates on phylogenetic trees with a node support index of more than 75 and a high level of nucleotide sequence similarity (98.5–100.0% for different genes). Phylogenetic lineages and sublineages were designated according to the classification accepted in the literature [32, 35‒40].
The study protocol was approved by the Local Ethical Committee of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor) (Protocol No. 6 of 24.03.2021).
Results
Characterization of the spectrum of G[P]-genotypes of RVA in Nizhny Novgorod
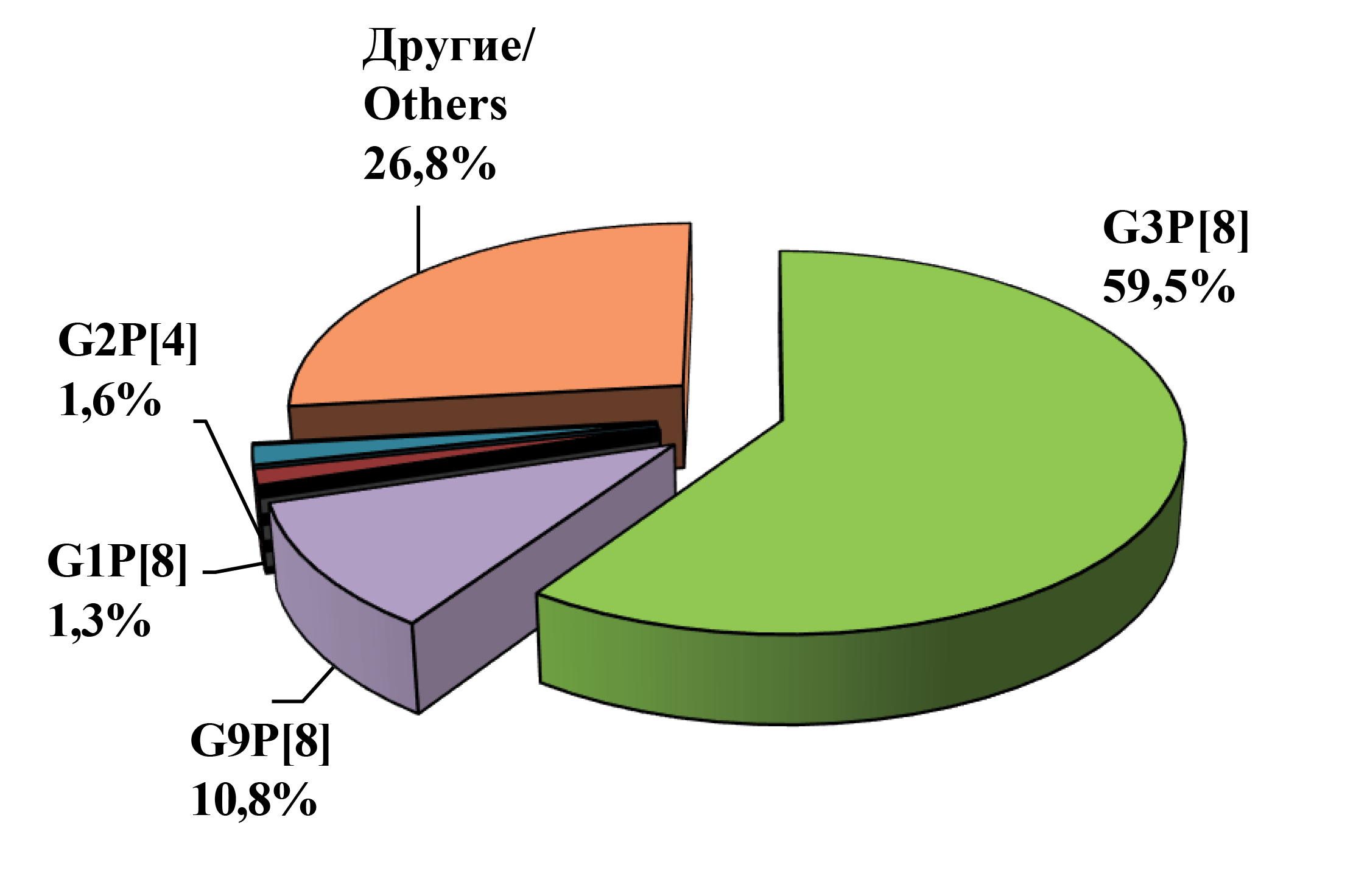
A total of 3715 stool samples from children hospitalized in an infectious disease hospital with symptoms of acute gastroenteritis between 2021 and 2023 were examined for the presence of rotavirus RNA. In 1085 cases (29.2%), RV RNA was detected, which was further used for G/[P]-genotyping by RT-PCR and/or sequencing. In 792 samples (72.9%), the G[P]-genotype of RV was determined. In 192 samples (17.7%), the G- or P-genotype was partially determined. The results of genotyping carried out on two genes (VP4 and VP7) showed that RVAs circulating in Nizhny Novgorod were characterized by high diversity. The spectrum of RV G-genotypes identified by PCR included 4 widely spread in the world (G1, G2, G3 and G9), and 2 rare for Russia (G6 and G8). The set of P-genotypes included 4 types, among which were frequently occurring P[4], P[8], and rare P[6] and P[9]. A total of 11 G/P combinations were detected: widespread G3P[8], G9P[8], G2P[4], G1P[8], and rare G8P[8], G3P[9], G9P[4], G3P[6], G2P[8], G6P[9] and G3P[4] (Fig. 1).
Fig. 1. Distribution of RVA strains of the main G[P]-genotypes in Nizhny Novgorod in the season 2021‒2023 (in %).
Рис. 1. Распределение штаммов РВА основных G[P]-генотипов в Нижнем Новгороде в сезон 2021‒2023 гг. (в %).
The dominant position during the study period was occupied by strains of the G3P[8] genotype (59.5%), followed by strains of the G9P[8] genotype (10.8%). The other genotypes showed low share contribution: G9P[4] – 2.1%, G1P[8] – 1.3%, G2P[4] – 1.6%, G3P[4] – 1.3%, G8P[8] – 1.1%, G3P[6] – 0.8%, G3P[9] – 1.0%, G2P[8] – 0.8%, G6P[9] – 0.1%.
Identification of reassortant rotaviruses and determination of their complete genotypes
Examination of RVA-containing samples by RNA-PAGE revealed 6 strains (2 strains of genotype G2P[4] and 4 strains of genotype G3P[9]) that exhibited so-called broad (AU-1-like) genomic segment migration profiles in PAGE (slowly migrating segment 5, rapidly migrating segments 6 and 11) (Fig. 2).
Fig. 2. Migration profiles of dsRNA segments of typical representatives of DS-1-/Wa-like rotaviruses and studied strains of genotypes G2P[4] and G3P[9] in PAGE. * − an unusual position of the segment relative to typical representatives of DS- and Wa-like gene groups.
Рис. 2. Профили миграции сегментов днРНК типичных представителей DS-1-/Wa-подобных ротавирусов и исследуемых штаммов генотипов G2P[4] и G3P[9] в ПААГ. * − необычное положение сегмента относительно типичных представителей DS- и Wa-подобных геногрупп.
Strains were sequenced for 11 genes (VP1–VP4, VP6, VP7, NSP1–NSP5/6) to determine their complete genotypes: G2-P[4]-I2-R2-C2-M2-A3-N2-T3-E2-H3, G2-P[4]-I2-R2-C2-M2-A3-N2-T3-E3-H2 и G3-P[9]-I2-R2-C2-M2-A3-N2-T3-E3-H3. The studied strains had a mixed set of genes from two different genetic groups, DS-1-like (genotypes numbered 2) and AU-1-like (genotypes numbered 3), indicating their reassortant origin.
Phylogenetic analysis of rotaviruses based on all genome segments
The nucleotide sequences of 11 genes were used to study the phylogenetic relationships of the 6 identified RV genotypes G2P[4] and G3P[9]. Phylogenetic analysis allowed us to study the intragenotype diversity of the studied strains at the lineage and sublineage level. The adapted trees are shown in Fig. 3, 4.
Fig. 3. Phylogenetic trees based on nucleotide sequences structural genes (VP1–VP4, VP6, VP7) of rotavirus strains A. ■ − strains obtained in this work; □ − Nizhny Novgorod strains retrieved from GenBank.
Рис. 3. Филогенетическое дерево, построенное на основе нуклеотидных последовательностей структурных генов (VP1–VP4, VP6, VP7) штаммов ротавируса А. ■ − штаммы, полученные в данной работе; □ − нижегородские штаммы, взятые из GenBank.
Fig. 4. Phylogenetic trees based on nucleotide sequences: nonstructural genes (NSP1–NSP5/6) of rotavirus strains A. ■ − strains obtained in this work; □ − Nizhny Novgorod strains retrieved from GenBank.
Рис. 4. Филогенетическое дерево, построенное на основе нуклеотидных последовательностей неструктурных генов (NSP1–NSP5/6) штаммов ротавируса А. ■ − штаммы, полученные в данной работе; □ − нижегородские штаммы, взятые из GenBank.
For each gene, the studied strains were included in 1 or 2 clusters. The genes VP4, VP7, NSP4 and NSP5/6 showed the highest diversity (Table).
Table. Subgenotype-level genomic constellations of rotaviruses from Nizhny Novgorod
Таблица. Геномные констелляции ротавирусов Нижнего Новгорода на уровне субгенотипов
№ | Strains Штаммы | Genome segments / Сегменты генома | ||||||||||
VP7 | VP4 | VP6 | VP1 | VP2 | VP3 | NSP1 | NSP2 | NSP3 | NSP4 | NSP5/6 | ||
Feline-like strains / Штаммы, подобные РВ кошек | ||||||||||||
1 | ВА222 | G3-3-e | P[9] | I2-XIV | R2-XIII | C2-IX | M2-XI | A3 | N1 | T3 | E2-XVII | H3 |
2 | NN1061/16 | G6-I | P[9] | I2-XIV | R2-XIII | C2-IX | M2-XI | A3 | N2-XVI | T3 | E2-XVII | H3 |
3 | NN148/17 | G3-3-e | P[9] | I2-XIV | R2-XIII | C2-IX | M2-XI | A3 | N2-XVI | T3 | E3 | H3 |
4 | NN2748/18 | G3-3-e | P[9] | I2-XIV | R2-XIII | C2-IX | M2-XI | A3 | N2-XVI | T3 | E3 | H3 |
Typical DS-1-like strains / Типичные Ds-1-подобные штаммы | ||||||||||||
5 | NN96/18 | G2-IVa-3 | P[4]-IV-b | I2-V-1 | R2-V-1 | C2-IVa-1 | M2-VII | A2-IVa-1 | N2-V-1 | T2-V-1 | E2-VI | H2-IVa-1 |
6 | NN425/18 | G2-IVa-3 | P[4]-IV-b | I2-V-1 | R2-V-1 | C2-IVa-1 | M2-VII | A2-IVa-1 | N2-V-1 | T2-V-1 | E2-VI | H2-IVa-1 |
7 | NN437/18 | G2-IVa-1 | P[4]-IV-b | I2-V-1 | R2-V-1 | C2-IVa-1 | M2-VII | A2-IVa-1 | N2-V-1 | T2-V-1 | E2-VI | H2-IVa-1 |
8 | NN560/18 | G2-IVa-2 | P[4]-IV-a | I2-V-2 | R2-V-2 | C2-IVa-2 | M2-V | A2-IVa-2 | N2-V-2 | T2-V-2 | E2-VII | H2-IVa-2 |
Strains obtained in this work / Штаммы, полученные в данной работе | ||||||||||||
9 | 2853/21 | G3-3-e | P[9] | I2-XIV | R2-XIII | C2-IX | M2-XI | A3 | N2-XVI | T3 | E3 | H3 |
10 | 2885/21 | G3-3-e | P[9] | I2-XIV | R2-XIII | C2-IX | M2-XI | A3 | N2-XVI | T3 | E3 | H3 |
11 | 347/22 | G3-3-e | P[9] | I2-XIV | R2-XIII | C2-IX | M2-XI | A3 | N2-XVI | T3 | E3 | H3 |
12 | 2619/22 | G3-3-e | P[9] | I2-XIV | R2-XIII | C2-IX | M2-XI | A3 | N2-XVI | T3 | E3 | H3 |
13 | 1473/21 | G2-IVa-3 | P[4]-IV | I2-XIV | R2-XIII | C2-IX | M2-XI | A3 | N2-XVI | T3 | E2-XVII | H3 |
14 | 2924/21 | G2-IVa-3 | P[4]-IV | I2-XIV | R2-XIII | C2-IX | M2-XI | A3 | N2-XVI | T3 | E3 | H2-IVa |
Note. The studied strains are highlighted in bold. DS-1-like alleles are marked in red, alleles of RV cats are blue, and Wa-like alleles are green.
Примечание. Исследуемые штаммы выделены жирным шрифтом. Красным цветом отмечены DS-1-подобные аллели, голубым – аллели РВ кошек, зеленым – Wa-подобные аллели.
Strains similar to feline rotavirus BA222
The 4 strains of genotype G3P[9] identified in this study belonged to a group of RVs similar to strain VA222 isolated from a cat. These strains (2853/21, 2885/21, 347/22, and 2619/21) were similar in 9 genes (VP1–VP4, VP6, VP7, NSP1, NSP3, NSP5/6) to the prototypical RV strain VA222, but differed from it in the NSP2 and NSP4 genes, which possessed other genotypes. The highest level of nucleotide sequence similarity was found for the VP2, VP3, VP7, NSP1, NSP3, and NSP5/6 genes and amounted to 98.7–100.0%. A lower level of homology was shown for VP1, VP4 and VP6 genes, ranging from 96.3–98.1%.
Also, the studied strains were related to the Nizhny Novgorod VA222-like RV genotypes G3P[9] (NN148/17, NN2748/18) and G6P[9] (NN1061-16) detected earlier, in 2016–2018. With isolates of genotype G3P[9] (NN148/17 and NN2748/18), the tested strains had a high level of nucleotide sequence homology for all 11 genes (99.1–100.0%). The strain of genotype G6P[9] (NN1061/16) showed the closest affinity for 8 genes (VP1–VP4, NSP1–NSP3, NSP5/6) with a level of nucleotide sequence similarity of 98.7–100.0%. A lower level of similarity was established for the VP6 gene (96.0%). The VP7 and NSP4 genes had a different genotype.
Atypical strains of the G2P[4] genotype
The two RVA strains of genotype G2P[4] (1473/21 and 2924/21) identified in the present work had a mixed set of genes. They carried a VA222-like base, while the VP4, VP7 genes and in one case the NSP5/6 gene were obtained as a result of reassortment from typical representatives of the DS-1-like genetic group.
The most closely related by divergent genes (VP4, VP7 and in one case NSP5/6) were strains of genotype G2P[4] from Nizhny Novgorod identified earlier in 2018 (NN560/18, NN96/18 and NN425/18). The level of nucleotide sequence similarity was quite high at 98.7–99.9%.
The rest of the genes (VP1–VP3, VP6, NSP1–NSP5/6) were closely related to the VA222-like group of RVs. Directly with prototypic VA222, samples 1473/21 and 2924/21 were most related to the prototypic VA222 by 3 genes: VP2, NSP3, NSP4 and VP2, VP3, NSP3, respectively. The nucleotide sequence similarity in the case of these genes reached 98.6–99.8%, while for the other genes it was lower, 95.2–97.6%.
The relatedness of the studied strains of genotype G2P[4] with the Nizhny Novgorod RVs of genotypes G3P[9] and G6P[9] (NN1061/16, NN148/17, NN2748/18), identified earlier, was also established for all VA222-like genes. Thus, the studied strain 1473/21 shared the highest number of genes (8 out of 11) with strain NN148/17 (G3P[9]) (VP1–VP3, VP6, NSP1–NSP3, NSP5/6), showing high nucleotide sequence similarity, 99.1–99.9%. It shared 7 genes (VP1–VP3, NSP2–NSP5/6) with strain NN1061/16 (G6P[9]) with a nucleotide sequence similarity level of 98.6–99.8%. Five genes were related to those of strain NN2748/18 (G3P[9]) (VP1, VP2, VP6, NSP2, NSP3). The homology of their nucleotide sequence was estimated at 98.7–99.7%.
Similarly, strain2924/21 had close phylogenetic relatedness with strain NN148/17 (G3P[9]) for 8 genes (VP1–VP3, VP6, NSP1–NSP4), showing high nucleotide sequence similarity of 98.8–99.8%. With strain NN2748/18 (G3P[9]), it shared 6 genes (VP1–VP3, VP6, NSP2, NSP3) with a homology level of 98.5–99.5%. In the case of strain NN1061/16 (G6P[9]), it showed close affinity for 5 genes (VP1–VP3, NSP2, NSP3) with a high level of nucleotide sequence similarity, 99.1–99.8%.
Discussion
This paper characterizes the diversity of RVA genotypes circulating in Nizhny Novgorod in 2021–2023. The spectrum is represented by 11 types dominated by G3P[8] (59.5%), followed by G9P[8] (10.8%). By June 2021, there was a decrease in the proportion of G9P[8] genotypes in the Nizhny Novgorod RV population, with a subsequent change of the dominant genotype to G3P[8] in July 2021. [41]. The data obtained serve as an addition to the information on genetic rearrangements in the RV population during their long-term circulation in Nizhny Novgorod, observations of which have been carried out since 1984 [13, 26, 32, 40–44].
The presented results also supplement the available data on the genotype structure of RV populations in Russia. According to the data of the reference center of monitoring for acute intestinal infections, in the winter-spring period of 2021 in the territory of the Russian Federation (Moscow, Tomsk, Irkutsk, Sverdlovsk, Novosibirsk regions, the Republic of Dagestan, Khabarovsk Territory and Kamchatka Territory), the dominance of the G9P[8] genotype remained, but a gradual increase in the proportion of the G3P[8] genotype was already observed. In 2022, there was a significant decrease in the frequency of occurrence of the G9P[8] genotype and the prevalence of the G3P[8] genotype was shown to prevail in the RV circulation in the territory of the Russian Federation, which remained in 2023. [4, 5]. This information is consistent with the data obtained in the present study.
RVs of the G3 genotype of the VP7 gene have a wide host range and are found in most susceptible animal species (including humans, rabbits, monkeys, pigs, birds, cats, dogs, horses, mice, cows and lambs) [7, 45, 46]. In human RV, it is mostly associated with the P[8] genotype of the VP4 gene, but is less frequently found in combination with the P[9] genotype [47–49]. The G3P[9] combination is prevalent mainly in RVs of cats and dogs [50], but due to reassortment, strains similar to animal RVs can occur in humans [51]. RVs of genotype G3P[9] were first detected in humans in 1982 in Japan and Israel [17, 22, 52]. Later, their appearance was registered in Thailand and Spain [53–59]. In the territory of Nizhny Novgorod, G3P[9] strains have been sporadically observed in the population since 1984. In early studies, they were studied on the basis of single genes (VP4, VP6, VP7, NSP4) [60, 61]; the first study of these strains on the basis of all genome segments was conducted in 2023 [40].
G3P[9] RVs are sporadically found in humans, forming a separate genetic group, isolated from Wa-like and DS-1-like viruses [49, 59, 62–64]. For human G3P[9], the possibility of at least 8 different combinations of complete genotypes has been shown: G3-P[9]-I3-R3-C3-M3-A3-N3-N3-T3-E3-H3 [9, 17], G3-P[9]-I3-R3-C3-M3-A3-N3-T3-E3-H6 [16, 22, 51], G3-P[9]-I2-R2-C2-M2-A3-N2-T1-E2-H3 [46, 52], G3-P[9]-I2-R2-C2-M2-A3-N1-T6-E2-H3, G3-P[9]-I2-R2-C2-C2-M2-A2-A3-N2-N2-T6-E2-H3 [65], G3-P[9]-I2-R2-C2-C2-M2-A2-A3-N1-T3-E2-H3 [66, 67], G3-P[9]-I3-R3-C2-C2-M3-A3-N1-T6-E3-H3 [68, 69], and G3-P[9]-I2-R2-C2-M2-A2-A3-N2-T3-E3-H3 [68, 70]. Interestingly, the NSP1 gene of all G3P[9] strains of animal and human origin invariably possesses the A3 genotype, whereas the genotype of the other genes can vary (VP6 – I2/I3; VP1 – R2/R3; VP2 – C2/C3; VP3 – M2/M3; NSP2 – N1/N2/N3; NSP3 – T1/T3/T6; NSP4 – E2/E3; NSP5/6 – H3/H6).
The G3P[9] RVAs identified and investigated in the present study possessed a combination of G3-P[9]-I2-R2-C2-M2-M2-A3-N2-T3-E3-H3 genotypes. Phylogenetic analysis showed that they shared 9 genes (VP1–VP4, VP6, VP7, NSP1, NSP3, NSP5/6) with feline RV VA222 (Italy, 2005). Out of all of these, VP2, VP3, VP7, NSP1, NSP3, and NSP5 genes had a high percentage of nucleotide sequence similarity (98.7–100.0%), while VP1, VP4, and VP6 had a lower percentage (96.3–98.1%). The Nizhny Novgorod strains differed from type VA222 in the NSP2 and NSP4 genes, which possessed other genotypes.
The results obtained show close relatedness of the studied RVs with earlier strains of genotype G3P[9] from Nizhny Novgorod. In the period 2016–2018, 3 similar RVA strains of genotype G3P[9] [40] were identified and studied on the basis of all genome segments. Their nucleotide sequences were used for phylogenetic analysis in the present work. The similarity of Nizhny Novgorod G3P[9] strains of different years amounted to 99.7–100.0% for different genes.
Intergroup reassortants based on the VA222-like combination were previously found in South Korea. S. Jeong et al. (2014) investigated RVA of genotype G3P[9] isolated in 2012 from an unvaccinated 9-year-old girl with symptoms of severe gastroenteritis. The strain studied had the genotype G3-P[9]-I2-R2-C2-C2-M2-M2-A2-A3-N2-T3-E3-H3. Phylogenetic analysis showed that it shared 9 genes (VP1–VP4, VP6, VP7, NSP1, NSP3, NSP5/6) with prototype VA222 strain, with different levels of nucleotide sequence similarity. Clustering of the studied strains with typical DS-1-like human RVs was established for the NSP2 gene, and for NSP4 – with an Australian strain of cat and dog RVs. This strain had a complex evolutionary origin, potentially involving reassortment events between cat RVs and DS-1-like human RVs [70].
Similar data were obtained by a group of researchers from China. M. Cao et al. examined 2 strains of G3P[9] genotype isolated in 2020 and 2023 from a 12-month-old girl and a 16-month-old boy, respectively. Both strains had the combination of genotypes: G3-P[9]-I2-R2-C2-M2-M2-A3-N2-T3-E3-H3. Phylogenetic analysis established that both studied RVAs shared 8 genes (VP1–VP4, VP7, NSP1, NSP3, and NSP4) with RVAs similar to cat and dog RVAs, with varying levels of nucleotide sequence similarity (99.3–99.6%). The remaining 3 genes (VP6, NSP2, and NSP5/6) were associated with typical DS-1-like human RVs (99.3–100.0%) [68].
This study shows the circulation of unusual strains that had the genotypes G2-P[4]-I2-R2-C2-C2-M2-A2-A3-N2-T3-E2-H3 and G2-P[4]-I2-R2-C2-C2-M2-A2-A3-N2-T3-E3-H2. The VP1‒VP3, VP6, NSP1, NSP3‒NSP5/6 genes of one of them and VP1–VP3, VP6, NSP1, NSP3, NSP4 genes of the other were related to the prototype strain VA222 of RVA (Italy, 2005). The remaining genes (namely VP4, VP7 and in the case of one NSP5/6 strain) originated from human DS-1-like strains of genotype G2P[4]. Strains with this combination of genotypes were identified and characterized by sequencing all genome segments for the first time. In Russia and in the world, no reports on the detection of similar G2P[4] strains have been previously noted. In all genes, both BA222 and DS-1-like, these strains had close relatives from Nizhny Novgorod, which suggests their local origin.
The reassortant strains characterized in this study could not be detected by G/[P]-genotyping by PCR, which is routinely used to study RV. This emphasizes the importance of using a full-genome classification system and modern sequencing methods to monitor the circulation of this pathogen.
Conclusion
The results of the study in combination with previously obtained data expand the understanding of the significant genetic diversity of RV and the role of reassortants in its maintenance. This information is important for the development of new rotavirus vaccines, understanding of evolutionary processes in the RVA population, and indicates the relevance of studying the genetic evolution of rare and novel virus strains. To control the emergence of new variants, continuous molecular monitoring for circulating RVA using full genomic classification is a necessity.
Funding. The study was conducted as part of the State Task of the Federal Service for Supervision of Consumer Rights Protection and Human Well-Being (Rospotrebnadzor).
Conflict of interest. The authors declare no apparent or potential conflicts of interest related to the publication of this article.
Ethics approval. The study was conducted with the informed consent of the patients. The research protocol was approved by the Local Ethics Committee of the FSBI «Academician I.N. Blokhina Nizhny Novgorod Scientific Research Institute of Epidemiology and Microbiology» of the Federal Service for Supervision of Consumer Right Protection and Human Welfare (Rospotrebnadzor) (Protocol No. 6 dated 24.03.2021).
About the authors
Elena I. Velikzhanina
Academician I.N. Blokhina Nizhny Novgorod Scientific Research Institute of Epidemiology and Microbiology of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
Author for correspondence.
Email: www.e_velikzhanina@mail.ru
ORCID iD: 0000-0003-4069-1427
Junior Researcher, laboratory of molecular epidemiology of viral infections
Russian Federation, 603950, Nizhny NovgorodTatiana A. Sashina
Academician I.N. Blokhina Nizhny Novgorod Scientific Research Institute of Epidemiology and Microbiology of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
Email: tatyana.sashina@gmail.com
ORCID iD: 0000-0003-3203-7863
PhD, Senior Researcher, laboratory of molecular epidemiology of viral infections
Russian Federation, 603950, Nizhny NovgorodOlga V. Morozova
Academician I.N. Blokhina Nizhny Novgorod Scientific Research Institute of Epidemiology and Microbiology of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
Email: Olga.morozova.bsc@gmail.com
ORCID iD: 0000-0002-8058-8187
PhD, Senior Researcher, laboratory of molecular epidemiology of viral infections
Russian Federation, 603950, Nizhny NovgorodAlexander Yu. Kashnikov
Academician I.N. Blokhina Nizhny Novgorod Scientific Research Institute of Epidemiology and Microbiology of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
Email: a.kashn@yandex.ru
ORCID iD: 0000-0003-1033-7347
Research assistant, laboratory of molecular epidemiology of viral infections
Russian Federation, 603950, Nizhny NovgorodNatalia V. Epifanova
Academician I.N. Blokhina Nizhny Novgorod Scientific Research Institute of Epidemiology and Microbiology of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
Email: epifanovanv@mail.ru
ORCID iD: 0000-0001-7679-8029
PhD, Leading Researcher, laboratory of molecular epidemiology of viral infections
Russian Federation, 603950, Nizhny NovgorodNadezhda A. Novikova
Academician I.N. Blokhina Nizhny Novgorod Scientific Research Institute of Epidemiology and Microbiology of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
Email: novikova_na@mail.ru
ORCID iD: 0000-0002-3710-6648
Professor. Head of the laboratory of molecular epidemiology of viral infections
Russian Federation, 603950, Nizhny NovgorodReferences
- GBD 2016 Diarrheal Disease Collaborators. Estimates of the global, regional, and national morbidity, mortality, and etiologies of diarrhea in 195 countries: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018; 18(11): 1211–28. https://doi.org/10.1016/S1473-3099(18)30362-1
- Troeger С., Khalil I.A., Rao P.C., Cao S., Blacker B.F., Ahmed T., et al. Rotavirus vaccination and the global burden of rotavirus diarrhea among children younger than 5 years. JAMA Pediatr. 2018; 172(10): 958–65. https://doi.org/10.1001/jamapediatrics.2018.1960
- Sindhu K.N., Babji S., Ganesan S. Impact of rotavirus vaccines in low and middle-income countries. Curr. Opin. Infect. Dis. 2017; 30(5): 473–81. https://doi.org/10.1097/QCO.0000000000000397
- State Report «On the state of sanitary and epidemiological welfare of the population in the Russian Federation in 2022». Moscow; 2023. (in Russian)
- State report «On the state of sanitary and epidemiological welfare of the population in the Russian Federation in 2023». Moscow; 2024. (in Russian)
- Rixon F., Taylor P., Desselberger U. Rotavirus RVA segments sized by electron microscopy. J. Gen. Virol. 1984; 56(1): 233–9. https://doi.org/10.1099/0022-1317-65-1-233
- Estes M.K., Kapikian A.Z. Rotaviruses. In: Fields B.N., Knipe D.M., Howley P.M., eds. Fields Virology. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2007: 1917–73.
- Estes M.K., Greenberg H.B. Rotaviruses. In: Knipe D.M., Howley P.M. eds. Fields Virology. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2013: 1347–1401.
- Matthijnssens J., Ciarlet M., Rahman M., Attoui H., Banyai K., Estes M.K., et al. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch. Virol. 2008; 153(8): 1621–9. https://doi.org/10.1007/s00705-008-0155-1
- RCWG. Rotavirus classification working group; 2024. Available at: https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg
- Matthijnssens J., Heylen E., Zeller M., Rahman M., Lemey P., Van Ranst M. Phylodynamic analyses of rotavirus genotypes G9 and G12 underscore their potential for swift global spread. Mol. Biol. Evol. 2010; 27(10): 2431–6. https://doi.org/10.1093/molbev/msq137
- Maes P., Matthijnssens J., Rahman M., Van Ranst M. Rota C: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol. 2009; 9: 238. https://doi.org/10.1186/1471 2180-9-238
- Sashina T.A., Morozova O.V., Epifanova N.V., Novikova N.A. Predominance of new G9P[8] rotaviruses closely related to Turkish strains in Nizhny Novgorod (Russia). Arch. Virol. 2017; 162(8): 2387–92. https://doi.org/10.1007/s00705-017-3364-7
- Kiseleva V., Faizuloev E., Meskina E., Marova A., Oksanich A., Samartseva T., et al. Molecular-genetic characterization of human rotavirus A strains circulating in Moscow, Russia (2009–2014). Virol. Sin. 2018; 33(4): 304–13. https://doi.org/10.1007/s12250-018-0043-0
- Doro R., Laszlo B., Martella V., Leshem E., Gentsch J., Parashar U., et al. Review of global rotavirus strain prevalence data from six years post vaccine licensure surveillance: is there evidence of strain selection from vaccine pressure? Infect. Genet. Evol. 2014; 28: 446–61. https://doi.org/10.1016/j.meegid.2014.08.017
- Santos N., Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 2005; 15(1): 29–56. https://doi.org/10.1002/rmv.448
- Matthijnssens J., Ciarlet M., Heiman E., Arijs I., Delbeke T., Mcdonald S.M., et al. Full genome-based classification of rotaviruses reveals a common origin between human Wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 2008; 82(7): 3204–19. https://doi.org/10.1128/JVI.02257-07
- Heiman E.M., Mcdonald S.M., Barro M., Taraporewala Z.F., Bar-Magen T. Ndpatton J.T. Group A human rotavirus genomics: evidence that gene constellations are influenced by viral protein interactions. J. Virol. 2008; 82(22): 11106–16. https://doi.org/10.1128/JVI.01402-08
- Uprety T., Wang D., Li F. Recent advances in rotavirus reverse genetics and its utilization in basic research and vaccine development. Arch. Virol. 2021; 166(9): 2369–86. https://doi.org/10.1007/s00705-021-05142-7
- Nakagomi O., Ohshima A., Aboudy Y., Shif I., Mochizuki M., Nakagomi T., et al. Molecular identification by RNA-RNA hybridization of a human rotavirus that is closely related to rotaviruses of feline and canine origin. J. Clin. Microbiol. 1990; 28(6): 1198–203. https://doi.org/10.1128/jcm.28.6.1198-1203.1990
- Matthijnssens J., Van Ranst M. Genotype constellation and evolution of group A rotaviruses infecting humans. Curr. Opin. Virol. 2012; 2(4): 426–33. https://doi.org/10.1016/j.coviro.2012.04.007
- Nakagomi T., Nakagomi O. RNA-RNA hybridization identifies a human rotavirus that is genetically related to feline rotavirus. J. Virol. 1989; 63(3): 1431–4. https://doi.org/10.1128/JVI.63.3.1431-1434.1989
- Tsugawa T., Rainwater-Lovett K., Tsutsumi H. Human G3P[9] rotavirus strains possessing an identical genotype constellation to AU-1 isolated at high prevalence in Brazil, 1997–1999. J. Gen. Virol. 2015; 96(Pt. 3): 590–600. https://doi.org/10.1099/vir.0.071373-0
- Cook N., Bridger J., Kendall K., Gomara M.I., El-Attar L., Gray J. The zoonotic potential of rotavirus. J. Infect. 2004; 48(4): 289–302. https://doi.org/10.1016/j.jinf.2004.01.018
- Jain S., Vashistt J., Changotra H. Rotaviruses: is their surveillance needed? Vaccine. 2014; 32(27): 3367–78. https://doi.org/10.1016/j.vaccine.2014.04.037 Novikova N.A., Sashina T.A., Epifanova N.V., Kashnikov A.U., Morozova O.V. Long-term monitoring of G1P[8] rotaviruses circulating without vaccine pressure in Nizhny Novgorod, Russia, 1984–2019. Arch. Virol. 2020; 165(4): 865–75. https://doi.org/10.1007/s00705-020-04553-2
- Gouvea V., Glass R.I., Woods P., Taniguchi K., Clark H.F., Forrester B., et al. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 1990; 28(2): 276–82. https://doi.org/10.1128/jcm.28.2.276-282.1990
- Gentsch J.R., Glass R.I., Woods P., Gouvea V., Gorziglia M., Flores J., et al. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 1992; 30(6): 1365–73. https://doi.org/10.1128/jcm.30.6.1365-1373.1992
- Maunula L., von Bonsdorff C.H. Short sequences define genetic lineages: phylogenetic analysis of group A rotaviruses based on partial sequences of genome segments 4 and 9. J. Gen. Virol. 1998; 79(Pt. 2): 321–32. https://doi.org/10.1099/0022-1317-79-2-321
- Iturriza-Gómara M., Isherwood B., Desselberger U., Gray J. Reassortment in vivo: driving force for diversity of human rotavirus strains isolated in the United Kingdom between 1995 and 1999. J. Virol. 2001; 75(8): 3696–705. https://doi.org/10.1128/JVI.75.8.3696-3705.2001
- Iturriza-Gómara M., Kang G., Gray J. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J. Clin. Virol. 2004; 31(4): 259–65. https://doi.org/10.1016/j.jcv.2004.04.009
- Sashina T.A., Morozova O.V., Epifanova N.V., Novikova N.A. Genotype constellations of the rotavirus A strains circulating in Nizhny Novgorod, Russia, 2017-2018. Infect. Genet. Evol. 2020; 85: 104578. https://doi.org/10.1016/j.meegid.2020.104578
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980; 16(2): 111–20. https://doi.org/10.1007/BF01731581
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018; 35(6): 1547–9. https://doi.org/10.1093/molbev/msy096
- Rahman M., Matthijnssens J., Yang X., Delbeke T., Arijs I., Taniguchi K., et al. Evolutionary history and global spread of the emerging g12 human rotaviruses. J. Virol. 2007; 81(5): 2382–90. https://doi.org/10.1128/JVI.01622-06
- Mukherjee A., Dutta D., Ghosh S., Bagchi P., Chattopadhyay S., Nagashima S., et al. Full genomic analysis of a human group A rotavirus G9P[6] strain from Eastern India provides evidence for porcine-to-human interspecies transmission. Arch. Virol. 2009; 154(5): 733–46. https://doi.org/10.1007/s00705-009-0363-3
- Ndze V.N., Esona M.D., Achidi E.A., Gonsu K.H., Doro R., Marton S., et al. Full genome characterization of human Rotavirus A strains isolated in Cameroon, 2010-2011: diverse combinations of the G and P genes and lack of reassortment of the backbone genes. Infect. Genet. Evol. 2014; 28: 537–60. https://doi.org/10.1016/j.meegid.2014.10.009
- Wang Y.H., Pang B.B., Ghosh S., Zhou X., Shintani T., Urushibara N., et al. Molecular epidemiology and genetic evolution of the whole genome of G3P[8] human rotavirus in Wuhan, China, from 2000 through 2013. PLoS One. 2014; 9(3): e88850. https://doi.org/10.1371/journal.pone.0088850
- Agbemabiese C.A., Nakagomi T., Doan Y.H., Nakagomi O. Whole genomic constellation of the first human G8 rotavirus strain detected in Japan. Infect. Genet. Evol. 2015; 35: 184–93. https://doi.org/10.1016/j.meegid.2015.07.033
- Sashina T.A., Velikzhanina E.I., Morozova O.V., Epifanova N.V., Novikova N.A. Detection and full-genotype characterization of rare and reassortant Rotavirus A strains in Nizhny Novgorod, European part of Russia. Arch. Virol. 2023; 168(8): 215. https://doi.org/10.1007/s00705-023-05838-y
- Morozova O.V., Sashina T.A., Epifanova N.V., Velikzhanina E.I., Novikova N.A. Phylodynamic characteristics of reassortant DS-1-like G3P[8]-strains of rotavirus type A isolated in Nizhny Novgorod (Russia). Braz. J. Microbiol. 2023; 54(4): 2867–77. https://doi.org/10.1007/s42770-023-01155-3
- Novikova N.A., Sashina T.A., Solntsev L.A., Epifanova N.V., Kashnikov A.Yu., Pogodina L.V., et al. Manifestations of epidemic process of rotavirus infection in Nizhny Novgorod in pre-vaccination period. Zhurnal mikrobiologii, epidemiologii i immunobiologii. 2017; 94(5): 46–52. https://doi.org/10.36233/0372-9311-2017-5-46-52 https://elibrary.ru/ysqaeq (in Russian)
- Morozova O.V., Sashina T.A., Novikova N.A. Detection and molecular characterization of reassortant DS-1-like G1P[8] strains of rotavirus A. Voprosy virusologii. 2017; 62(2): 91–6. https://doi.org/10.18821/0507-4088-2017-62-2-91-96 https://elibrary.ru/yjkhhh
- Sashina T.A., Morozova O.V., Epifanova N.V., Kashnikov A.U., Leonov A.V., Novikova N.A. Molecular monitoring of the rotavirus (Reoviridae: Sedoreovirinae: Rotavirus: Rotavirus A) strains circulating in Nizhny Novgorod (2012–2020): detection of the strains with the new genetic features. Voprosy virusologii. 2021; 66(2): 140–51. https://doi.org/10.36233/0507-4088-46 https://elibrary.ru/azvpec (in Russian)
- Martella V., Ciarlet M., Camarda A., Pratelli A., Tempesta M., Greco G. et al. Molecular characterization of the VP4, VP6, VP7, and NSP4 genes of lapine rotaviruses identified in Italy: emergence of a novel VP4 genotype. Virology. 2003; 314(1): 358–70. https://doi.org/10.1016/s0042-6822(03)00418-5
- Grant L., Esona M., Gentsch J., Watt J., Reid R., Weatherholtz R., et al. Detection of G3P[3] and G3P[9] rotavirus strains in American Indian children with evidence of gene reassortment between human and animal rotaviruses. J. Med. Virol. 2011; 83(7): 1288–99. https://doi.org/10.1002/jmv.22076
- Nakagomi O., Kaga E. Distinctness of NSP1 gene of human rotavirus AU-1 from NSP1 gene of other human genogroups. Res. Virol. 1995; 146(6): 423–8. https://doi.org/10.1016/0923-2516(96)80902-2
- Theamboonlers A., Veravigrom M., Yambangyang O., Trairatvorakul P., Chongsrisawat V., Poovorawan Y. The incidence of rotavirus a isolates of G genotype in Thailand in 2002–2004. Acta Virol. 2005; 49(2): 111–5.
- Martella V.A., Potgieter C., Lorusso E., De Grazia S., Giammanco G.M., Matthijnssens J., et al. A feline rotavirus G3P[9] carries traces of multiple reassortment events and resembles rare human G3P[9] rotaviruses. J. Gen. Virol. 2011; 92(Pt. 5): 1214–21. https://doi.org/10.1099/vir.0.027425-0
- Matthijnssens J., Ciarlet M., Mcdonald S.M., Attoui H., Banyai K., Brister J.R. et al. Uniformity of rotavirus strain nomenclature proposed by the rotavirus classification working group (RCWG). Arch. Virol. 2011; 156(8): 1397–413. https://doi.org/10.1007/s00705-011-1006-z
- Theamboonlers A., Maiklang O., Thongmee T., Chieochansin T., Vuthitanachot V., Poovorawan Y. Complete genome analysis of a rare human G3P[9] rotavirus posing as an AU-1 like strain. Springerplus. 2013; 2: 569. https://doi.org/10.1186/2193-1801-2-569
- Wang Y.H., Pang B.B., Zhou X., Ghosh S., Tang W.F. Peng J.S., et al. Complex evolutionary patterns of two rare human G3P[9] rotavirus strains possessing a feline/canine-like H6 genotype on an AU-1-like genotype constellation. Infect. Genet. Evol. 2013; 16: 103–12. https://doi.org/10.1016/j.meegid.2013.01.016
- Sanchez-Fauquier A., Montero V., Moreno S., Sole M., Colomina J., Iturriza-Gomara M., et al. Human rotavirus G9 and G3 as major cause of diarrhea in hospitalized children, Spain. Emerg. Infect. Dis. 2006; 12(10): 1536–41. https://doi.org/10.3201/eid1210.060384
- Khamrin P., Maneekarn N., Peerakome S., Tonusin S., Phan T.G., Okitsu S., et al. Molecular characterization of rare G3P[9] rotavirus strains isolated from children hospitalized with acute gastroenteritis. J. Med. Virol. 2007; 79(6): 843–51. https://doi.org/10.1002/jmv.20840
- Inoue Y., Kitahori Y. Rare group a rotavirus G3P[9] isolated in Nara Prefecture, Japan. Jpn J. Infect. Dis. 2006; 59(2): 139–40.
- Iizuka M., Chiba M., Masamune O., Kaga E., Nakagomi T., Nakagomi O. A highly conserved genomic RNA constellation of Japanese isolates of human rotaviruses carrying G serotype 3 and P serotype 9. Res. Virol. 1994; 145(1): 21–4. https://doi.org/10.1016/s0923-2516(07)80003-3
- Griffin D.D., Nakagomi T., Hoshino Y., Nakagomi O., Kirkwood C.D., Parashar U.D., et al. Characterization of nontypeable rotavirus strains from the United States: identification of a new rotavirus reassortant (P2A[6], G12) and rare P3[9] strains related to bovine rotaviruses. Virology. 2002; 294(2): 256–69. https://doi.org/10.1006/viro.2001.1333
- Gollop R., Nakagomi O., Silberstein I., Shulman L.M., Greenberg H.B., Mendelson E., et al. Three forms of AU-1 like human rotaviruses differentiated by their overall genomic constellation and by the sequence of their VP8*. Arch. Virol. 1998; 143(2): 263–77. https://doi.org/10.1007/s007050050285
- Kaga E., Iizuka M., Nakagomi T., Nakagomi O. The distribution of G (VP7) and P (VP4) serotypes among human rotaviruses recovered from Japanese children with diarrhea. Microbiol. Immunol. 1994; 38(4): 317–20. https://doi.org/10.1111/j.1348-0421.1994.tb01784.x
- Sashina T.A., Morozova O.V., Novikova N.A. G/[P]-types of rotavirus A in Nizhny Novgorod: 2012-2014. Infektsiya i immunitet. 2014; 4(1): 91. https://elibrary.ru/vzqyan (in Russian)
- Novikova N.A., Ponomareva N.V., Novikov D.V., Prilipov A.G., Epifanova N.V., Golitsyna L.N. Nucleotide sequence analysis of the nsp4 gene from group a rotavirus-es isolated in Nizhni Novgorod. Voprosy virusologii. 2008; 53(6): 35–9. https://elibrary.ru/kaxfub (in Russian)
- Nakagomi O., Nakagomi T., Oyamada H., Suto T. Relative frequency of human rotavirus subgroups 1 and 2 in Japanese children with acute gastroenteritis. J. Med. Virol. 1985; 17(1): 29–34. https://doi.org/10.1002/jmv.1890170105
- Nakagomi O., Nakagomi T. Molecular evidence for naturally occurring single VP7 gene substitution reassortant between human rotaviruses belonging to two different genogroups. Arch. Virol. 1991; 119(1-2): 67–81. https://doi.org/10.1007/BF01314324
- Nakagomi O., Nakagomi T. Molecular epidemiology of human rotaviruses: genogrouping by RNA–RNA hybridization. Arch. Virol. 1996; 12: 93–8. https://doi.org/10.1007/978-3-7091-6553-9_11
- De Grazia S., Giammanco G.M., Potgieter C.A., Matthijnssens J., Banyai K., Platia M.A., et al. Unusual assortment of segments in 2 rare human rotavirus genomes. Emerg. Infect. Dis. 2010; 16(5): 859–62. https://doi.org/10.3201/eid1605.091826
- Banyai K., Laszlo B., Duque J., Steele A.D., Nelson E. Anthony S., et al. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine. 2012; 30(1): 122–30. https://doi.org/10.1016/j.vaccine.2011.09.111
- Tsugawa T., Hoshino Y. Whole genome sequence and phylogenetic analyses reveal human rotavirus G3P[3] strains Ro1845 and HCR3A are examples of direct virion transmission of canine/feline rotaviruses to humans. Virology. 2008; 380(2): 344–53. https://doi.org/10.1016/j.virol.2008.07.041
- Cao M., Yuan F., Zhang W., Wang X., Ma J., Ma X., et al. Genomic analysis of two rare human G3P[9] rotavirus strains in Ningxia, China. Infect. Genet. Evol. 2023; 116: 105518. https://doi.org/10.1016/j.meegid.2023.105518
- Mijatovic-Rustempasic S., Roy S., Sturgeon M., Rungsrisuriyachai K., Esona M.D., Degroat D., et al. Full-genome sequence of a rare human G3P[9] rotavirus strain. Genome Announc. 2014; 2(2): e00143–14. https://doi.org/10.1128/genomeA.00143-14
- Jeong S., Lim I., Kim W. Whole-genome analysis of a rare human Korean G3P[9] rotavirus strain suggests a complex evolutionary origin potentially in evolving reassortment events between feline and bovine rotaviruses. PLoS One. 2014; 9(5): e97127. https://doi.org/10.1371/journal.pone.0097127
Supplementary files