Analysis of genetic polymorphisms and drug resistance mutations in the NS5 region of HCV genome (Flasuviricetes: Amarillovirales: Flaviviridae: Hepacivirus C) in samples obtained in 2022–2023 from HIV-infected treatment-naive residents of Altai Krai
- Authors: Lapovok I.A.1, Syrkina A.V.1, Kirichenko A.A.1, Shlykova A.V.1, Lukyanenko N.V.2, Safyanova T.V.2, Safronova A.E.2, Shevchenko V.V.2,3, Kireev D.E.1
-
Affiliations:
- Central Research Institute of Epidemiology
- Altai State Medical University (ASMU)
- Altai Regional Center for Prevention and Control of AIDS and Infectious Diseases
- Issue: Vol 70, No 3 (2025)
- Pages: 224-233
- Section: ORIGINAL RESEARCHES
- URL: https://virusjour.crie.ru/jour/article/view/16722
- DOI: https://doi.org/10.36233/0507-4088-298
- EDN: https://elibrary.ru/NZBCOE
- ID: 16722
Cite item
Abstract
Introduction. Altai Krai is a region with an unfavorable situation of HIV-1 and HCV infection, as well as HIV-1 and HCV coinfection. Due to this, it is necessary to study the HCV genetic variants and their drug resistance (DR) to direct-acting antivirals (DAAs) in patients with HIV-1 and HCV coinfection.
Aim of the study. The analysis of HCV genome fragments encoding NS5A and NS5B proteins in samples obtained from treatment-naïve residents of Altai Krai with newly diagnosed HIV and HCV co-infection to determine the genetic variant of HCV and genetic features of the virus associated with its sensitivity to NS5A and NS5B inhibitors.
Materials and methods. Blood plasma samples (n = 286) collected in 2022–2023 from HIV-infected individuals were analyzed for HCV markers. The HCV RNA concentration was measured, nucleotide sequences of NS5A and NS5B and Core (for HCV 2k/1b samples) fragments were obtained, the subtype was determined, and DR and polymorphic positions were analyzed.
Results. Antibodies to HCV were detected in 94/286 (32.86%) samples, sequences were obtained from 52 samples. Subtypes 3a, 1b, recombinant form 2k/1b and subtype 1a were found in 28 (53.85%), 17 (32.69%), 5 (9.62%) and one (1.92%) samples, respectively. One sample harbored HCV 1b + 3a mix-infection. Reduced sensitivity (5.66%) and complete resistance (9.43%) to the NS5A inhibitor daclatasvir were most often detected. Certain gene polymorphisms were identified in the sequences.
Conclusion. Our results may indirectly indicate the increasing proportion of the HCV subtype 3a in the hepatitis C epidemic in the Altai Territory. Our data on DR and polymorphisms should be taken into account in antiviral therapy of patients.
Keywords
Full Text
Introduction
By the early 2020s, chronic hepatitis C virus (HCV) infection had been detected in more than 70 million people worldwide, and about 15 million people in Europe [1]. According to the World Health Organization (WHO), in 2022, approximately 242,000 people worldwide lost their lives to the consequences of HCV infection1.
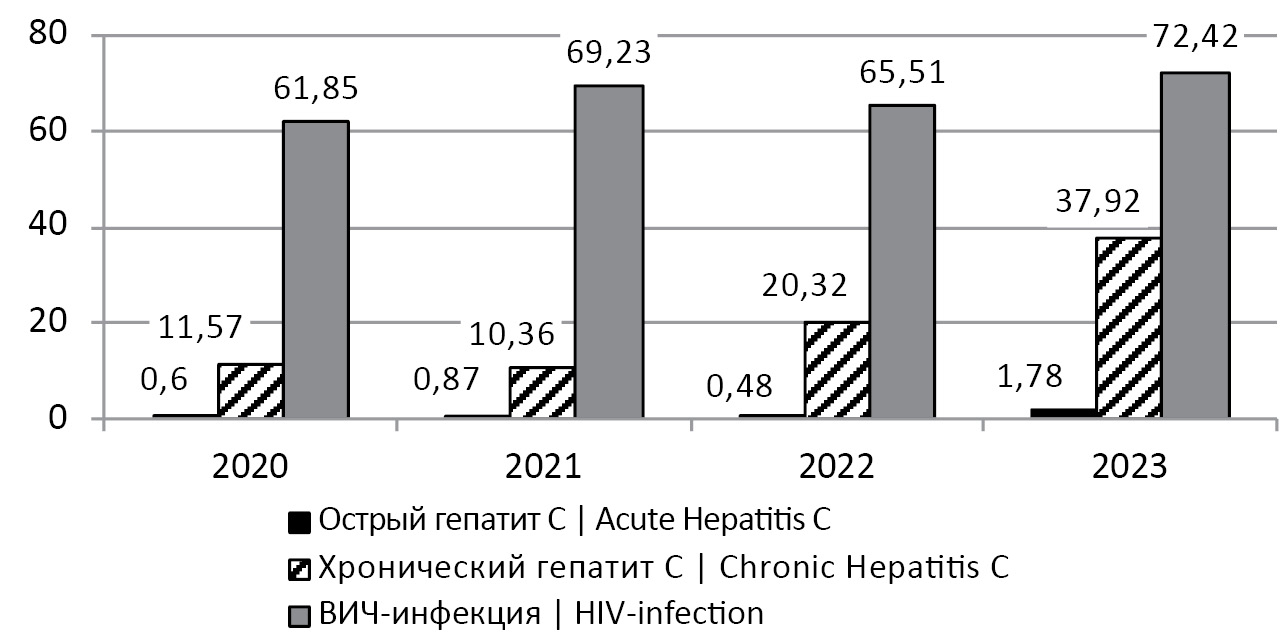
The problem of the spread of such infections as HCV and human immunodeficiency virus (HIV-1) is severe both in Russia as a whole and in certain regions of the country. In recent years, Altai Krai has been classified as an HIV-unfavorable region. Thus, the incidence of HIV infection in the period 2020–2023 did not fall below 61 cases per 100 thousand population, reaching a record high of 72.42 cases per 100 thousand population in 2023 (Fig. 1)2,3,4,5. At the same time, more than 65% of HIV-infected persons detected in Altai Krai from 1990 to 2023 were over 30 years old, i.e. belonged to the socially and economically active population6.
Fig. 1. Incidence (per 100 thousand population) of acute, chronic hepatitis C and HIV infection in Altai Krai in 2020–2023.
Рис. 1. Заболеваемость (на 100 тыс. населения) острым, хроническим гепатитом C и ВИЧ-инфекцией в Алтайском крае в 2020–2023 гг.
The established insignificant decrease in the incidence of hepatitis C and HIV infection in Altai Krai in 2021–2022 is probably related to the COVID-19 epidemic, in which the self-isolation regime did not allow effective detection of new HIV-1 and HCV infections. In Europe as a whole, there was a decrease in the incidence of HCV infection in 2020–2021: from 6.6 cases per 100,000 population in 2019 to 4.7 per 100,000 population by 2021. However, this rate once again increased to 6.5 per 100,000 population in 2022 [1]. Thus, and in Altai Krai, an increase in the incidence of acute and chronic hepatitis C was registered in 2023: up to 1.78 and 37.92 per 100 thousand population, respectively (Fig. 1).
The complexity of HIV-1 and HCV (HIV-1/HCV) co-infection is due to the fact that both infections can exacerbate the course of one another. Curing the HIV infection, with the exception of certain unique cases, is impossible, requiring lifelong therapy. Regarding HCV infection, there are cases where acute hepatitis is eradicated spontaneously, the frequency of which reaches, according to certain data, 18–34% [2], according to other data – 15–45% [3]. In the remaining cases, chronic infection develops, which can lead to liver cirrhosis, hepatocellular carcinoma (HCC) and liver failure, which is especially dangerous during antiretroviral therapy (ART) of HIV-1 [3, 4]. As a result, patients with HIV-1/HCV co-infection are more than 5 times more likely to develop severe liver disease compared to HCV mono-infection [5]. Co-infected patients have a 2-fold higher risk of developing liver cirrhosis [6] compared to HCV mono-infected patients. Finally, co-infected patients are more likely to develop chronic kidney disease (by almost 50%) and acute renal failure (by almost 64%) than patients with HIV infection alone [7]. As a result, mortality in HCV/HIV-1 co-infection is higher than in mono-infection [3, 5].
Currently, 8 genotypes and more than 100 subtypes of HCV have been registered [8], but the number of known subtypes of the virus is constantly increasing. HCV subtypes 1b, 1a and 3a are the most frequently recorded HCV infections in Russia [4]. Subtype 1b is the dominant variant. It accounted for about 55% of HCV infections in Russia in 2016 [9]. As of November 1, 2024. 46.1% and 36.2% of all nucleotide sequences of the HCV genome from Russia (n = 1593) uploaded to the international database of the Los Alamos Institute, USA (https://hcv.lanl.gov/) belonged to subtypes 1b and 3a, respectively.
The distribution of HCV genotypes in Russia is heterogeneous. For example, a 2022 study analyzing HCV in 35 regions of Russia showed that in the country as a whole, HCV genotype 1 accounted for 53.6% of infections, and genotype 3 accounted for 35.4%. At the same time, in the Far East of the country, 60 and 39% of HCV infections were caused by virus of genotypes 1 and 3, respectively; the ratio of these subtypes was 58%/35% in the Central Federal District, 52%/41% in the Northwestern Federal District, 53%/38% in the Southern Federal District, 58%/34% in the North Caucasus Federal District, 59%/35% in the Volga Federal District, and 57%/35% in the Ural Federal District. In the Siberian Federal District, which includes Altai Krai, HCV infection with genotype 1 and 3 accounted for 50 and 43% of cases, respectively [10].
In addition to the described HCV genotypes, the recombinant form 2k/1b, first described in 2002 in St. Petersburg, is also circulating in the country. It likely emerged in the USSR in the period of 1923–1956. The recombination point between 2k and 1b is located in the NS2 region of the viral genome, thus all genome regions encoding structural proteins of 2k/1b belong to the subtype 2k and regions encoding non-structural proteins belong to subtype 1b [11, 12].
In 2016, the WHO program for the elimination of HCV in the world by 2030 was developed. According to it, measures should be taken to reduce the number of new HCV cases by 90% and mortality by 65%. At the same time, the percentage of detected HCV-infected persons should reach 90%, and the percentage of cured persons – up to 80%7 [1, 3].
A therapy regimen including pegylated interferon (peg-IFN) and ribavirin has been the standard of care for chronic hepatitis C patients for many years [4]. However, since 2011, direct-acting antivirals (DAAs) aimed at inhibiting viral non-structural proteins NS3/4A (a protein combining the functions of protease and helicase), NS5A (a multifunctional protein involved in viral replication and assembly) and NS5B (RNA-dependent RNA polymerase) have been used to treat HCV infection [2–4]. The use of DAAs has achieved sustained virologic success in more than 90% of chronic hepatitis C therapies while reducing the duration of treatment to 12 weeks [3, 9, 13].
Despite the fact that pangenotypic DAAs universal for all HCV variants dominating in the world have been introduced into clinical practice [8], the determination of the virus genotype remains an urgent task. HCV genotype determines both the course of infection and the strategy of HCV infection therapy. Thus, subtype 1b is less sensitive to interferon-based drugs; therefore, the use of DAAs is recommended for HCV 1b therapy [9]. Moreover, only 50% of cases when peg-IFN-α and ribavirin were used to treat HCV genotypes 1 and 4 resulted in sustained virologic success, while side effects of such therapy were observed in at least 10% of patients. HCV subtype 3a is sensitive to interferon therapy, and HCV subtype 1a is more likely to develop drug resistance (DR) to DAAs than HCV subtype 1b [3].
Before the introduction of DAAs into widespread practice, the treatment of HCV infection caused by the 2k/1b genetic variant remained a separate problem. Upon being incorrectly genotyped as a genotype 2 virus, peg-IFN and ribavirin-based therapy, to which 2k/1b has a reduced sensitivity, as does subtype 1b virus, could be prescribed for treatment of the patient [11]. Although current HCV treatment recommendations do not include this regimen, there is a necessity to adequately differentiate 2k/1b from other viral variants when investigating the genetic diversity and epidemiology of HCV.
The widespread use of DAAs inevitably leads to the problem of DR, including transmissible resistance, i.e., viral resistance in patients without experience with therapy. A 2022 study in the North Caucasus Federal District showed that 5/42 (12%) patients without therapy experience had HCV with mutations to at least one DAA drug [4]. At the same time, different HCV genotypes, as a rule, differ in the frequency of occurrence of certain DR mutations.
The most significant from the clinical point of view is DR to NS5A protein inhibitors, since it is the target of first-line therapy drugs [9]. Nucleotide substitutions in at least 12 positions of NS5A associated with resistance to inhibitors of this protein are well known [2]. The most important and frequently encountered in the world are substitutions in positions 28, 30, 31 and 93 [14]. It is also the same case for Russia. A 2018 study showed that among HCV subtype 1b variants circulating in Moscow in 2008–2014, DR to NS5A inhibitors was found with a frequency of more than 22%, with the most frequently detected substitutions R30Q, L31M and Y93H in the NS5A protein [9]. The presence of combinations of L31F + Y93H or Y93H + A62S/T + A30K substitutions may be a prognostic sign of ineffectiveness of daclatasvir therapy in HCV genotype 3 [13, 14]. And the presence of S98G + Y93H combination in NS5A can double the resistance of genotype 3 HCV to daclatasvir compared to HCV containing only Y93H [15]. Meanwhile, the failure of therapy with NS5A inhibitors has also been observed in patients with HCV infection and combination of S98G mutation with A30K or A62T substitution [4, 15].
The combined use of NS5A inhibitors together with NS5B inhibitors has proved to be excellent. For example, concomitant use of NS5A inhibitors daclatasvir or velpatasvir together with NS5B inhibitor sofosbuvir is included in the Hepatitis C Control Program in Pakistan, the country with the second highest HCV prevalence in the world [13].
There are 24 known mutations of HCV resistance to NS5B inhibitors [2]. Analysis of HCV NS5B fragment from Russian patients from the Northwestern Federal District with newly diagnosed HIV infection in 2020 showed that the D310N substitution (an unfavorable marker of HCV infection progression to liver pathology) was rarely detected among HCV subtype 3a samples, and the C316N substitution associated with low-level resistance to tegobuvir was detected among almost 30% of subtype 1b viruses [3]. The E237G substitution can reduce sensitivity to various NS5B inhibitors [4].
In addition, a number of NS5B polymorphisms have been described whose presence is not investigated in the evaluation of DR, but which have the potential to influence the efficacy of therapy. A polymorphism of a genetic variant is defined as a substitution at a position in the genome (compared to a reference sequence) that is common in more than 1% of samples of that variant. The combination of D148N + I363V, A150V + I363V and T227S + S183P NS5B polymorphisms can reduce the sensitivity of subtype 3a virus to ribavirin by 1.3–1.6 times [16], while the A150V substitution in HCV genotype 3 can reduce the sensitivity to interferon-α by more than 12 times [17].
The aim of the study was the analysis of HCV genome fragments encoding NS5A and NS5B proteins in samples obtained from treatment-naïve residents of Altai Krai with newly diagnosed HIV and HCV co-infection to determine the genetic variant of HCV and genetic features of the virus associated with its sensitivity to NS5A and NS5B inhibitors.
Materials and methods
In 2022–2023, a collection of blood plasma samples from AIDS Center patients with newly diagnosed HIV infection without experience of specific antiviral therapy was collected at the Altai Krai Center for the Prevention and Control of AIDS and Infectious Diseases (AIDS Center) as part of routine analysis of HIV-1 and HCV infection.
The study was approved by the Ethics Committee of the Central Research Institute of Epidemiology (protocol No. 93 of 18.06.2019).
The presence of antibodies to HCV (anti-HCV) was determined by enzyme immunoassay using the commercially available «Best anti-HCV» kit (Vector Best, Novosibirsk, Russia).
HCV RNA concentration (viral load, VL) in blood plasma samples was measured using the AmpliSens HCV-Monitor-FL reagent kit (Central Research Institute of Epidemiology, Moscow, Russia).
Subsequent RNA extraction, amplification and sequencing of NS5A (1 to 117 amino acids) and NS5B fragments (148 to 556 amino acids) were performed using the AmpliSens HCV-Resist-Seq reagent kit (Central Research Institute of Epidemiology, Moscow, Russia) and the Applied Biosystems 3500 genetic analyzer (LifeTechnologies, USA).
For the obtained nucleotide sequences, we determined the genetic variant of HCV and DR using the HCVBlast online tool of the information resource of the Los Alamos Institute (USA) [18] and the geno2pheno[HCV] online tool of the information portal of the Genophore non-profit scientific society [19]. Interpretation of the degree of DR was performed using the geno2pheno[HCV] online tool: the virus is resistant (in the case of the presence of a well-characterized mutation associated with DR) or has a reduced sensitivity (in the case of the presence of a mutation presumed to be associated with resistance but for which insufficient evidence of clinical outcome has been collected) [19].
For samples assigned to the recombinant form of HCV 2k/1b on the basis of NS5A- and NS5B-fragment analysis, additional sequencing of the Core fragment of the HCV genome (8 to 191 amino acids) was performed using the in-laboratory technique and Sanger sequencing [20], followed by Core nucleotide sequence analysis using HCVBlast [18].
Nucleotide sequences were analyzed for polymorphisms using the MEGA 6.0 program [21]. NS5A and NS5B nucleotide positions described in the literature were analyzed [2, 4, 9, 13–17, 22].
Results
A total of 286 blood plasma samples were collected from HIV-infected individuals, 139 (48.60%) males and 147 (51.40%) females. The main route of HIV-1 infection for patients was sexual 247 (86.36%), for 39 (13.64%) – injection drug use (IDU). The vulnerable group of injection drug users (IDUs) was more often male, 31/39 (79.49%), while males accounted for only 46.96% (116/247) of those with sexual route of infection. The mean duration of infection from diagnosis to specimen collection was 15 days (95% confidence interval (CI) 12.42–18.76).
A total of 94/286 (32.86%) individuals were found to have HCV infection. Among the 94 patients with HIV-1/HCV infection, 57 (60.64%) were male. The mean age of patients was 41 years (95% CI 39.23–42.70). Co-infection was most commonly detected in males in the age group of 35–44 years and females in the age group of 55–64 years. A total of 34/94 (36,17%) patients with co-infection belonged to the vulnerable IDU group, while only 5/192 (2.60%) patients with HIV-1 mono-infection belonged to the vulnerable IDU group.
HCV VL < 150 IU/mL was present in 34/94 (36.17%) samples from co-infected patients, which did not allow further genetic analysis. The mean VL in the remaining 60 samples was 6.96 log10 (95% CI 6.75–7.11) IU/mL. Nucleotide sequences of NS5A and NS5B fragments of the HCV genome were obtained for 52/60 (86.67%) samples with HCV VL > 150 IU/mL.
Genotype analysis based on real-time polymerase chain reaction (PCR) and subsequent analysis with HCVBlast and geno2pheno[HCV] tools revealed that 28/52 (53.85%) samples had HCV that belonged to subtype 3a, 17 (32.69%) samples had HCV subtype 1b, 5 (9.62%) samples had HCV recombinant form 2k/1b, and one sample had HCV subtype 1a. HCV types 1a and 3a were detected in a sample obtained from a 25-year-old female patient from Barnaul with an IDU route of infection and duration of HIV infection of about 6 months, indicating co-infection with two HCV variants.
The recombinant nature of all 5 samples with HCV 2k/1b was confirmed by Core fragment analysis. Interestingly, all 5 2k/1b samples were attributed by NS5B to HCV subtype 1b by the geno2pheno[HCV] program, whereas similar NS5A analysis and analysis of both fragments in HCVBlast allowed unmistakable identification of the 2k/1b variant.
Since one sample had a co-infection with two HCV variants, 53 virus genomes were further analyzed. The result of DR analysis to NS5A and NS5B inhibitors using geno2pheno[HCV] tool is shown in Fig. 2. The drugs ombitasvir (NS5A inhibitor) and dasabuvir (NS5B inhibitor) are not recommended for therapy of HCV subtype 3a infection. Thus, DR to these drugs was assessed for 24 non-3a-subtype genomes.
Fig. 2. Result of drug resistance analysis to NS5A and NS5B inhibitors of all HCV nucleotide sequences (n = 53) and non-3a subtypes (n = 24).
Рис. 2. Результат анализа лекарственной устойчивости к ингибиторам NS5A и NS5B всех нуклеотидных последовательностей ВГС (n = 53) и не 3a-субтипов (n = 24).
The result of the analysis of DR mutations, polymorphisms and atypical mutations in DR-critical positions of the genomic fragments studied is shown in the Table.
Table. Results of HCV nucleotide sequence (n = 53) analysis for the presence of DR mutations and significant polymorphisms
Таблица. Результат анализа нуклеотидных последовательностей ВГС (n = 53) на наличие мутаций ЛУ и важных полиморфизмов
Substitution Замена | Subtype 3a Субтип 3a, abs./абс. (%) (n = 29) | Subtype 1b Субтип 1b, abs./абс. (%) (n = 17) | 2k/1b, abs./абс. (%) (n = 5) | Subtype 1a Субтип 1a, abs./абс. (%) (n = 2) | The drug affected by substitution Препарат, на который влияет замена |
NS5A fragment (aa. 1–117), abs. (%) Фрагмент NS5A (aa. 1–117), абс. (%) | |||||
M28V | – | – | – | 1 (50) | Velpatasvir, ombitasvir Велпатасвир, омбитасвир |
R30Q | – | 1 (5.88) | 1 (20) | – | Daclatasvir, ombitasvir Даклатасвир, омбитасвир |
R30H | – | 1 (5.88) | – | – | Daclatasvir Даклатасвир |
A30K | 3 (10.35) | – | – | – | Elbasvir, ledipasvir, pibrentasvir, velpatasvir, daclatasvir Элбасвир, ледипасвир, пибрентасвир, велпатасвир, даклатасвир |
A62L | 1 (3.45) | – | – | – | Daclatasvir Даклатасвир |
A30S/T | 2 (6.90) | – | – | – | |
L31M | – | 1 (5.88) | – | – | |
A62S | 22 (75.86) | – | – | – | |
A62T | 3 (10.35) | – | – | – | |
A62V | 2 (6.90) | – | – | – | |
A92V | – | – | 1 (20) | – | |
Y93H | – | – | – | – | |
S98G | 6 (20.70) | – | – | – | |
P587S | – | 1 (5.88) | – | – | |
NS5B fragment (aa. 148–556), abs. (%) Фрагмент NS5B (aa. 148–556), абс. (%) | |||||
L159F | – | 7 (41.18) | – | – | Sofosbuvir Софосбувир |
S556G | – | 3 (17.65) | – | – | Dasabuvir, sofosbuvir Дасабувир, софосбувир |
C316N | – | 9 (52.94) | – | – | Tegobuvir Тегобувир |
S368A | – | 1 (5.88) | – | – | |
N444D | – | – | – | 2 (100) | |
C451H/T/Y | – | 4 (23.53) | – | – | |
S556A | – | 1 (5.88) | – | – | |
D148N | – | – | – | – | |
A150V | 7 (24.14) | – | – | – | |
S183P | – | 17 (100) | 5 (100) | 2 (100) | |
T227S | – | – | – | – | |
E237G | – | – | – | – | |
D310N | 24 (82.76) | – | – | – | |
I363V | – | – | – | – | |
Reduced sensitivity or complete resistance were most frequently detected to the NS5A inhibitor daclatasvir: in 5.66% and 9.43% of viruses, respectively. This was associated with R30Q/H and L31M mutations in 3 sequences of subtype 1b and one sequence of 2k/1b, and A30K and A62L substitutions in 3 sequences and one sequence of subtype 3a, respectively. The A30K substitution was responsible for resistance to elbasvir and ledipasvir, as was the L31M substitution detected in one sequence of subtype 1b. It is also worth noting the high level of polymorphism at position 62 in subtype 3a viruses: 22 (75.86%) samples contained the A62S substitution, another 5 contained the A62T/V substitution, and only one contained the daclatasvir DR-related A62L substitution.
The two samples mentioned above (subtype 1b and 2k/1b) with the R30Q substitution and resistance to daclatasvir were also resistant to ombitasvir. Sequence of subtype 1a from a sample with co-infection with two HCV variants had reduced sensitivity to velpatasvir and ombitasvir due to the M28V mutation. However, no resistance was detected in subtype 3a sequence from the same patient.
Also 6 (20.69%) subtype 3a sequences contained S98G substitution. However, no associated Y93H mutation was detected in any sample.
In the NS5B fragment, DR mutations were detected only among subtype 1b sequences: the L159F and S556G mutations associated with DR to sofosbuvir contained 7 (41.18%) and 3 (17.65%) samples, respectively. Another 9 (52.94%) subtype 1b sequences contained C316N mutation associated with DR to tegobuvir.
The D310N polymorphism was detected in 24 (82.76%) subtype 3a sequences, and the A150V substitution was present in 7 (24.14%) sequences of this subtype.
As for the combinations of polymorphisms at positions 148, 183, 227 and 363 affecting the sensitivity of HCV subtype 3a to ribavirin, only S183P polymorphism was detected in all sequences of subtype 1b, 2k/1b and 1a. However, not a single sample of subtype 3a contained this substitution.
Finally, non-DR polymorphisms were found in the NS5B nucleotide sequences examined, but at positions critical for virus resistance to dasabuvir. A total of 4 (23.53%) subtype 1b viruses contained C451H/T/Y polymorphisms, and none contained an DR mutation at this C451S position. One 1b sample had an S368A substitution instead of S368T, and another had an S556A substitution instead of S556G/N/R. Both samples of subtype 1a had the N444D substitution instead of N444K [22].
Discussion
In the surveyed group of HIV-infected patients, only slightly more than 32% were infected with HCV. This is probably due to the fact that HIV-1/HCV co-infection is more common in IDUs: while the prevalence of HCV is 10–14% in individuals practicing high-risk sexual behavior, among IDUs this percentage is increased to 85–90% [5]. Since only 13.64% of patients in the study were IDUs, it is not surprising that less than 1/3 of them had HIV-1/HCV co-infection. Meanwhile, the proportion of IDUs among HIV-1/HCV-infected patients was significantly higher than in HIV-1-only patients: 36.17% vs. 2,60%.
Gender and age indicators of the surveyed group of patients with detected HIV-1/HCV co-infection were similar to those for HCV in Europe. Thus, in European countries, there are 1.6–1.9 cases per woman with HCV among males, and the mean age of infected individuals is about 46 years [1]. In the present study, the ratio of women to men with co-infection was 1 : 1.54, and the average age of patients was 41 years old.
We identified the predominance of HCV subtype 3a in the studied collection of samples. This result differs from the data of studies showing the distribution of HCV genotypes both in Russia as a whole and in the Siberian Federal District [9, 10], which can be regarded as a distinctive feature of the HCV epidemic among HIV-infected individuals in Altai Krai. The fact that HCV subtype 3a is prevalent in Altai Krai is alarming because, compared to other genotypes, infection with genotype 3 leads to a more rapid development of fibrosis and a high degree of hepatic steatosis, as well as to an increased risk of HCC [13]. Moreover, among all genotypes, genotype 3 most frequently demonstrates resistance to DAAs [15].
The problem of HCV subtyping is of great importance not only for the understanding the epidemiology of the pathogen, but also for predicting the course of infection and choosing effective therapy. To establish the recombinant form 2k/1b circulating in Russia, genotyping of the region to the left of NS2 (e.g., Core) and to the right of NS2 (NS3, NS5A or NS5B) is necessary. In recent years, therapy regimens that do not include interferon-based drugs, but also include pangenotypic DAAs, have been introduced into widespread practice. In Russia, not only regimens including sofosbuvir and daclatasvir, but also 2nd line therapy regimens containing such drugs as elbasvir, ledipasvir, ombitasvir and velpatasvir are widely used [23]. In this regard, the necessity to analyze the nucleotide sequence of the Core region is reduced. This leads to a complication in the differentiation of form 2k/1b from subtype 1b, since only the nucleotide sequence to the right of NS2 is often available for HCV samples. Our results of subtyping of NS5A and NS5B fragments suggest that the use of NS5A fragment analysis in the geno2pheno[HCV] program and/or NS5A or NS5B fragment analysis in the HCVBlast program is sufficient to establish 2k/1b. In this case, the fragments obtained as part of the HCV DR test for DAA drugs are suitable for analysis.
The case of infection with two HCV variants detected in this study has a logical explanation – the main route of HCV infection in the world and in Europe is currently IDU [1, 5, 8]. The patient with co-infection with two HCV subtypes belongs to the vulnerable group of IDUs. Simultaneous infection with several virus variants is another serious cause of HCV genetic variability, as it creates a basis for the emergence of recombinant forms of the virus [12]. In the case described here, there was the simultaneous presence of subtype 1a with reduced sensitivity to ombitasvir and subtype 3a, which requires a special approach to therapy. Potentially, this coinfection could lead to the formation of a recombinant with reduced sensitivity to individual therapy regimens.
The high frequency of the C316N substitution in subtype 1b samples (52.94% of samples) is consistent with previously published data on the high prevalence of this substitution among the 1b variant circulating in Russia [3]. This substitution is associated with low-level resistance to tegobuvir in HCV subtype 1b. At the same time, we did not identify the better known DR C316Y mutation associated with resistance to NS5B inhibitors in subtypes 1a and 1b [3, 22].
In more than 82% of subtype 3a sequences, we detected the D310N substitution associated with the progression of liver pathology in HCV. Previously, this substitution was detected in only 30% of subtype 3a sequences isolated in Russia [3]. The increase in its prevalence among this genetic variant is a cause for concern. At the same time, more than 24% of subtype 3a sequences contained the A150V substitution associated with significant resistance of the virus to interferon-α [17]. Thus, the HCV subtype 3a variant circulating among HIV-infected individuals in Altai Krai may not only lead to severe liver damage, but also be resistant to interferon-based therapy.
Meanwhile, the present study did not reveal any combination of NS5B polymorphisms in subtype 3a described in the literature [16], which can insignificantly affect the efficacy of ribavirin therapy. However, we found a number of polymorphisms in subtype 1b and 1a sequences that are not associated with DR, but present in critical nucleotide positions.
Conclusion
Thus, the results obtained in general indicate the predominance of HCV subtype 3a in recently detected HIV-infected individuals in Altai Krai and indirectly – an increase in the percentage of this genetic variant in the hepatitis C epidemic in the studied region of the country. Data on the identified mutations and genetic polymorphisms should be taken into account when prescribing antiviral therapy for patients.
1 WHO. Hepatitis C. Available at: https://who.int/news-room/fact-sheets/detail/hepatitis-c
2 The State report «On the state of sanitary and epidemiological welfare of the population in the Russian Federation in 2020». Moscow; 2021.
3 The State report «On the state of sanitary and epidemiological welfare of the population in the Russian Federation in 2021». Moscow; 2022.
4 The State report «On the state of sanitary and epidemiological welfare of the population in the Russian Federation in 2022». Moscow; 2023.
5 The State report «On the state of sanitary and epidemiological welfare of the population in the Russian Federation in 2023». Moscow; 2024.
6 Altai Regional Center for the Prevention and Control of AIDS and Infectious Diseases. Available at: https://altaids22.ru
7 WHO. Global health sector strategy on viral hepatitis, 2016–2021: towards ending viral hepatitis; 2016. Доступно по: http://apps.who.int/iris/bitstream/10665/246177/1/WHO-HIV-2016.06-eng.pdf
About the authors
Ilya A. Lapovok
Central Research Institute of Epidemiology
Author for correspondence.
Email: i_lapovok@mail.ru
ORCID iD: 0000-0002-6328-1415
Cand. Sci. (Biol.), Senior Researcher, HIV diagnostic and molecular epidemiology laboratory
Russian Federation, 111123, MoscowArina V. Syrkina
Central Research Institute of Epidemiology
Email: syrkina@cmd.su
ORCID iD: 0009-0003-2733-6663
Junior Researcher, HIV diagnostic and molecular epidemiology laboratory
Russian Federation, 111123, MoscowAlina A. Kirichenko
Central Research Institute of Epidemiology
Email: kirichenko@cmd.su
ORCID iD: 0000-0002-7116-0138
Cand. Sci. (Med.), Senior Researcher, HIV diagnostic and molecular epidemiology laboratory
Russian Federation, 111123, MoscowAnastasia V. Shlykova
Central Research Institute of Epidemiology
Email: murzakova_a.v@mail.ru
ORCID iD: 0000-0002-1390-8021
Researcher, HIV diagnostic and molecular epidemiology laboratory
Russian Federation, 111123, MoscowNatalya V. Lukyanenko
Altai State Medical University (ASMU)
Email: natvalluk@mail.ru
ORCID iD: 0000-0002-0003-5145
Dr. Sci. (Med.), Professor, Department of Epidemiology, Microbiology and Virology
Russian Federation, 656038, BarnaulTatyana V. Safyanova
Altai State Medical University (ASMU)
Email: tvsafyanova@yandex.ru
ORCID iD: 0000-0003-3293-4265
Dr. Sci. (Med.), Professor, Head of the Department of Epidemiology, Microbiology and Virology
Russian Federation, 656038, BarnaulArina E. Safronova
Altai State Medical University (ASMU)
Email: safariev00@mail.ru
ORCID iD: 0009-0009-7350-6073
Resident of the Department of Epidemiology, Microbiology and Virology
Russian Federation, 656038, BarnaulValery V. Shevchenko
Altai State Medical University (ASMU); Altai Regional Center for Prevention and Control of AIDS and Infectious Diseases
Email: infecgepatit@yandex.ru
ORCID iD: 0000-0001-6282-5495
Cand. Sci. (Med.), Chief Non-staff Infectiologist, Associate Professor, Department of Epidemiology, Microbiology and Virology
Russian Federation, 656038, Barnaul; 656010, BarnaulDmitry E. Kireev
Central Research Institute of Epidemiology
Email: dmitkireev@yandex.ru
ORCID iD: 0000-0002-7896-2379
Cand. Sci. (Biol.), Head, HIV diagnostic and molecular epidemiology laboratory
Russian Federation, 111123, MoscowReferences
- Simão M., Gonçalves C. Hepatitis C virus infection in Europe. Pathogens. 2024; 13(10): 841. https://doi.org/10.3390/pathogens13100841
- Martinez M.A., Franco S. Therapy implications of hepatitis C virus genetic diversity. Viruses. 2021; 13(1): 41. https://doi.org/10.3390/v13010041
- Ostankova Yu.V., Valutite D.E., Zueva E.B., Serikova E.N., Shchemelev A.N., Boumbaly S., et al. Primary HCV drug resistance mutations in patients with newly diagnosed HIV infection. Problemy osobo opasnykh infektsii. 2020; (3): 97–105. https://elibrary.ru/laroef (in Russian)
- Valutite D., Ostankova Y., Semenov A., Lyalina L., Totolian A. Distribution of primary resistance mutations in Saint Petersburg in patients with chronic hepatitis C. Diagnostics. 2022; 12(5): 1054. https://doi.org/10.3390/diagnostics12051054
- Sulkowski M.S. Viral hepatitis and HIV coinfection. J. Hepatol. 2008; 48(2): 353–67. https://doi.org/10.1016/j.jhep.2007.11.009
- Thein H.H., Yi Q., Dore G.J., Krahn M.D. Natural history of hepatitis C virus infection in HIV-infected individuals and the impact of HIV in the era of highly active antiretroviral therapy: a meta-analysis. AIDS. 2008; 22(15): 1979–91. https://doi.org/10.1097/QAD.0b013e32830e6d51
- Wyatt C.M., Malvestutto C., Coca S.G., Klotman P.E., Parikh C.R. The impact of hepatitis C virus coinfection on HIV-related kidney disease: a systematic review and meta-analysis. AIDS. 2008; 22(14): 1799–807. https://doi.org/10.1097/QAD.0b013e32830e0152
- Vo-Quang E., Pawlotsky J.M. «Unusual» HCV genotype subtypes: origin, distribution, sensitivity to direct-acting antiviral drugs and behaviour on antiviral treatment and retreatment. Gut. 2024; 73(9): 1570–82. https://doi.org/10.1136/gutjnl-2024-332177
- Kichatova V.S., Karlsen A.A., Isaeva O.V., Solonin S.A., Malinnikova E.Yu., Kyuregyan K.K., et al. Drug resistant variants of hepatitis C Virus genotype1B in Russia: analysis of aminoacid substitutions in NS5A and core proteins. Zhurnal infektologii. 2018; 10(4): 30–6. https://elibrary.ru/vvmeki (in Russian)
- Pimenov N., Kostyushev D., Komarova S., Fomicheva A., Urtikov A., Belaia O., et al. Epidemiology and genotype distribution of hepatitis C virus in Russia. Pathogens. 2022; 11(12): 1482. https://doi.org/10.3390/pathogens11121482
- Akimov I.A., Timofeev D.I., Mavzyutov A.R., Ivanov M.K. Detection of circulating HCV recombinant form rf1_2k/1b in blood serum of patients by real-time RT-PCR. Klinicheskaya laboratornaya diagnostika. 2021; 66(2): 122–8. https://elibrary.ru/vvobzb (in Russian)
- Kalinina O., Norder H., Mukomolov S., Magnius L.O. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 2002; 76(8): 4034–43. https://doi.org/10.1128/jvi.76.8.4034-4043.2002
- Mushtaq S., Hashmi A.H., Khan A., Asad Raza Kazmi S.M., Manzoor S. Emergence and Persistence of Resistance-Associated Substitutions in HCV GT3 Patients Failing Direct-Acting Antivirals. Front. Pharmacol. 2022; 13: 894460. https://doi.org/10.3389/fphar.2022.894460
- Wang C., Valera L., Jia L., Kirk M.J., Gao M., Fridell R.A. In vitro activity of daclatasvir on hepatitis C virus genotype 3 NS5A. Antimicrob. Agents. Chemother. 2013; 57(1): 611–3. https://doi.org/10.1128/AAC.01874-12
- Fernandes Campos G.R., Ward J., Chen S., Bittar C., Rodrigues J.P.V., de Lourdes Candolo Martinelli A., et al. A novel substitution in NS5A enhances the resistance of hepatitis C virus genotype 3 to daclatasvir. J. Gen. Virol. 2021; 102(1): jgv001496. https://doi.org/10.1099/jgv.0.001496
- Mejer N., Fahnøe U., Galli A., Ramirez S., Weiland O., Benfield T., et al. Mutations identified in the hepatitis C virus (HCV) polymerase of patients with chronic HCV treated with ribavirin cause resistance and affect viral replication fidelity. Antimicrob. Agents Chemother. 2020; 64(12): e01417–20. https://doi.org/10.1128/AAC.01417-20
- Lee W.Y.J., Jones M., Wing P.A.C., Rajagopal S., Foster G.R. The A150V polymorphism of genotype 3 hepatitis C virus polymerase inhibits interferon alfa by suppressing protein kinase R activation. Cell. Mol. Gastroenterol. Hepatol. 2021; 11(4): 1163–75. https://doi.org/10.1016/j.jcmgh.2020.11.012
- Los Alamos HCV database. Available at: https://hcv.lanl.gov
- Kalaghatgi P., Sikorski A.M., Knops E., Rupp D., Sierra S., Heger E., et al. Geno2pheno [HCV] – a web-based interpretation system to support hepatitis C treatment decisions in the era of direct-acting antiviral agents. PLoS One. 2016; 11(5): e0155869. https://doi.org/10.1371/journal.pone.0155869
- Kichatova V.S., Kyuregyan K.K., Soboleva N.V., Karlsen A.A., Isaeva O.V., Isaguliants M.G., et al. Frequency of interferon-resistance conferring substitutions in amino acid positions 70 and 91 of core protein of the Russian HCV 1b isolates analyzed in the T-cell epitopic context. J. Immunol. Res. 2018; 2018: 7685371. https://doi.org/10.1155/2018/7685371
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013; 30(12): 2725–9. https://doi.org/10.1093/molbev/mst197
- Kati W., Koev G., Irvin M., Beyer J., Liu Y., Krishnan P., et al. In vitro activity and resistance profile of dasabuvir, a nonnucleoside hepatitis C virus polymerase inhibitor. Antimicrob. Agents Chemother. 2015; 59(3): 1505–11. https://doi.org/10.1128/AAC.04619-14
- Valutite D.E., Semenov A.V., Ostankova Yu.V., Kozlov K.V., Borisov A.G., Nazarov V.D. et al. Detection of drug resistance mutations of hepatitis C virus in patients with failure of the treatment with direct acting antivirals. Zhurnal mikrobiologii, epidemiologii i immunobiologii. 2021; 98(1): 18–27. https://elibrary.ru/aksnkx (in Russian)
Supplementary files