Detection of the Liman tick virus (unclassified Chuviridae) in tick cell line HAE/CTVM8
- Authors: Litov A.G.1,2, Shchetinin A.M.3, Kholodilov I.S.1, Belova O.A.1, Kalyanova А.S.1, Gushchin V.A.3,4, Karganova G.G.1,2
-
Affiliations:
- Chumakov Federal Scientific Center for Research and Development of Immune-and-Biological Products of RAS (Institute of Poliomyelitis)
- Institute for Translational Medicine and Biotechnology, Sechenov University
- Gamaleya Federal Research Centre for Epidemiology and Microbiology, Ministry of Health of the Russian Federation
- Federal State Autonomous Educational Institution of Higher Education I.M. Sechenov First Moscow State Medical University of the Ministry of Health of the Russian Federation (Sechenov University)
- Issue: Vol 70, No 2 (2025)
- Pages: 147-153
- Section: ORIGINAL RESEARCHES
- URL: https://virusjour.crie.ru/jour/article/view/16705
- DOI: https://doi.org/10.36233/0507-4088-283
- EDN: https://elibrary.ru/hrnscv
- ID: 16705
Cite item
Abstract
Introduction. Tick cell lines are widely used to study the biology of ticks and tick-borne pathogens, especially viruses. Most of the cell cultures currently available have been obtained from tick embryonic cells and can be infected with viruses. The HAE/CTVM8 cell line was obtained from Hyalomma anatolicum ticks and is often used for isolation of novel viruses.
The aim of the work is to study the HAE/CTVM8 cell line using high-throughput sequencing in order to search for viruses in it.
Materials and methods. The HAE/CTVM8 cell culture fluid was ultracentrifuged. The resulting pellet was used for high-throughput sequencing after RNA extraction, reverse transcription reaction, and synthesis of the second strand. The resulting reads were filtered by length and quality in the Trimmomatic program, after which the contigs were assembled using the SPAdes program and analyzed for the presence of viral sequences. The final assembly of the virus genome was carried out in the Ugene program. Sequence alignment was performed by the MAFFT program. The phylogenetic trees were constructed using the IQ-TREE program.
Results. We have identified the persistence of one virus, Liman tick virus (LMTV), in HAE/CTVM8 cell culture. Phylogenetically LMTV belongs to the Chuviridae – novel family, that consists of viruses detected by high-throughput sequencing, the virological characteristics of which are currently unknown.
Conclusion. The obtained information is of significant importance when utilizing HAE/CTVM8 cell culture in scientific research and during the process of isolating new viruses. Our study shows that this cell line with persistent LMTV is a ready-to-use system for studying Chuviridae reproduction
Keywords
Full Text
Introduction
Ticks are vectors of dangerous diseases such as Lyme disease, tick-borne encephalitis and many others [1, 2]. In this regard, the study of biology, physiology of ticks and ways to control them is becoming increasingly important. Tick cell cultures have become an indispensable tool for researchers [3]. First of all, they are extremely useful for studying the physiology and genetics of ticks [3, 4]; however, they are much more often used to propagate tick-borne pathogenic viruses [5, 6] and bacteria [7].
Attempts to culture tick cells have been ongoing for more than 50 years. Early studies led to the production of primary cultures of tick cells and/or tissues capable of surviving up to 6 months [8]. Later, continuous cell lines of different tick species capable of long-term cultivation in laboratory conditions were obtained [3, 9].
Most of the currently available tick cell lines have been derived from embryonic cells. This specificity of obtaining cell cultures, of the tick virome, leads to the fact that viruses persist in many tick cell cultures [10–12]. For example, Orbivirus saintcroixense (St. Croix River virus) has been detected in IDE2 cell cultures derived from Ixodes scapularis ticks as well as RA243 and RA257 from Rhipicephalus appendiculatus ticks [10, 11]. Three rhabdoviruses, IRE/CTVM19-associated rhabdovirus, Chimay rhabdovirus and Norway mononegavirus 1, were detected immediately in IRE/CTVM19 cell culture [12]. Furthermore, screening of a large number of available tick cell cultures with pan-nairovirus oligonucleotides yielded positive results, although doubts remained as to whether the amplification was specific [10]. Thus, tick cell cultures themselves can be a source of novel viruses. Nevertheless, the use of cells with persistent viral infection may affect the results of scientific research, especially when isolating and studying the properties of other viral or bacterial intracellular agents.
Modern approaches to the discovery of new viruses involve the use of high-throughput sequencing in combination with bioinformatic methods, which allows the discovery of tens and hundreds of new viruses in the process [13–16]. This approach has proven to be extremely effective in characterizing new arthropod viruses [14, 16], including ticks [13, 15], and was also used earlier to characterize viruses persisting in IRE/CTVM19 cell culture [12].
One of the achievements of next generation sequencing was the recent isolation of a new family of (–)RNA-containing viruses, the Chuviridae. To date, this family includes 16 genera and 43 species of viruses that have been detected in spiders, crustaceans, insects, fish and reptiles. The largest genus of the family is the Mivirus genus, which includes 10 species that are mainly associated with different species of ixodid ticks. Unclassified chivirid или chuvirus-like sequences have been found in cephalopod mollusks, termites and turtles. The Chuviridae family is the largest in the Jingchuvirales order, which is related to the Mononegavirales order, which includes many well-known (–)RNA-containing viruses [17].
According to bioinformatic data, representatives of Chuviridae are viruses with single-stranded (–)RNA that encode 2 to 4 open reading frames, including polymerase, glycoprotein, and nucleoprotein. Genomes of different representatives of Chuviridae can be linear or circular, segmented or unsegmented [17].
HAE/CTVM8 cell culture was obtained from Hyalomma anatolicum ticks [9] and is routinely used for isolation and maintenance of various viruses [8, 18–20]. However, it remains unknown whether this culture contains any persistent viruses. There is only some evidence to suggest the presence of unknown nairo-like viruses in this cell culture [10].
The aim of this study is to research the HAE/CTVM8 cell line using next generation sequencing in order to search for viruses in it.
Materials and methods
HAE/CTVM8 cells were cultured in L-15 medium (Chumakov Federal Scientific Center for Research and Development of Immune-and-Biological Products of RAS (Institute of Poliomyelitis), Russia) with 10% tryptose-phosphate broth (Difco, USA), 20% fetal bovine serum (Gibco, USA), 2 mM L-glutamine and antibiotics, according to the previously described method [21]. The culture fluid was collected and clarified by centrifugation at 10,000 rpm for 30 min at +4 °C using a SW-28 rotor on an Optima L-90K Ultracentrifuge (Beckman Coulter, USA). After that, the supernatant was ultracentrifuged at 25,000 rpm for 6 h at +4 °C in the same rotor and centrifuge tube.
For high-throughput sequencing, the pellet after ultracentrifugation was dissolved in 200 μL PBS. RNA extraction was performed using the RNeasy Mini Kit (QIAGEN, Hilden, Germany). Reverse transcription reaction was performed with a random hexanucleotide primer using RevertAid (ThermoFisher Scientific, USA). Next, second strand synthesis was performed using NEBNext Ultra II Non-Directional RNA Second Strand Synthesis Module kit (NEB, USA). The resulting DNA was purified using Ampure XP (Beckman Coulter, USA). Libraries were prepared using the NEBNext Fast DNA Library Prep Set for Ion Torrent (NEB, USA) and sequenced on an Ion S5XL instrument using an Ion 530 Chip.
The obtained reads were filtered by length (at least 35 nt) and quality (Q20) using the Trimmomatic v.0.39 program [22]. The quality of the reads was checked using the FaQCs program [23]. The processed reads were assembled into contigs using the SPAdes v. 3.13.0 program using the option «−RNAviral». The obtained contigs were filtered from sequences with low complexity and non-viral sequences according to the previously described methodology [24]. In the remaining contigs, virus-containing contigs were searched according to the previously described methodology [24]. To improve the obtained assembly and cut off possible errors, we additionally performed read mapping using the Liman tick virus genome (GenBank number MN542376) as a reference genome in the Ugene v. 50.0 program [25].
For phylogenetic analysis, the complete amino acid sequences of the L protein (RNA polymerase of the Chuviridae family) for the identified virus, all complete sequences of the LMTV L protein available in the GenBank database, several selected representatives of Mivirus boleense, and representatives of genera and phylogenetic clades belonging to the Chuviridae family were obtained. Sequences were aligned with the MAFFT v. 7.310 program using the E-INS-i algorithm [26]. Uncertainly aligned regions were trimmed using the TrimAL v. 1.4. rev 15 program with automatic detection [27]. A phylogenetic tree was constructed using the maximum likelihood method in the program IQ-TREE v. 2.3.2 [28] with 1000 bootstrap replicates and automatic detection of the substitution model [29]. Phylogenetic trees were visualized in the FigTree v. 1.4.4 program. The genome was manually annotated and visualized using the GenomeDrawing tool (https://github.com/justNo4b/GenomeDrawing).
Results
In this study, high-throughput sequencing of the transcriptome of the culture fluid of a pure culture of HAE/CTVM8 cells derived from H. anatolicum ticks was performed. Filtering of reads by length and quality resulted in 2.2 million reads. Subsequent data processing revealed the presence of a single virus genome in the reads obtained. Preliminary analysis showed that this virus is genetically close to LMTV: 92.5% similarity in the nucleotide composition of the complete genome, according to the blastn program.
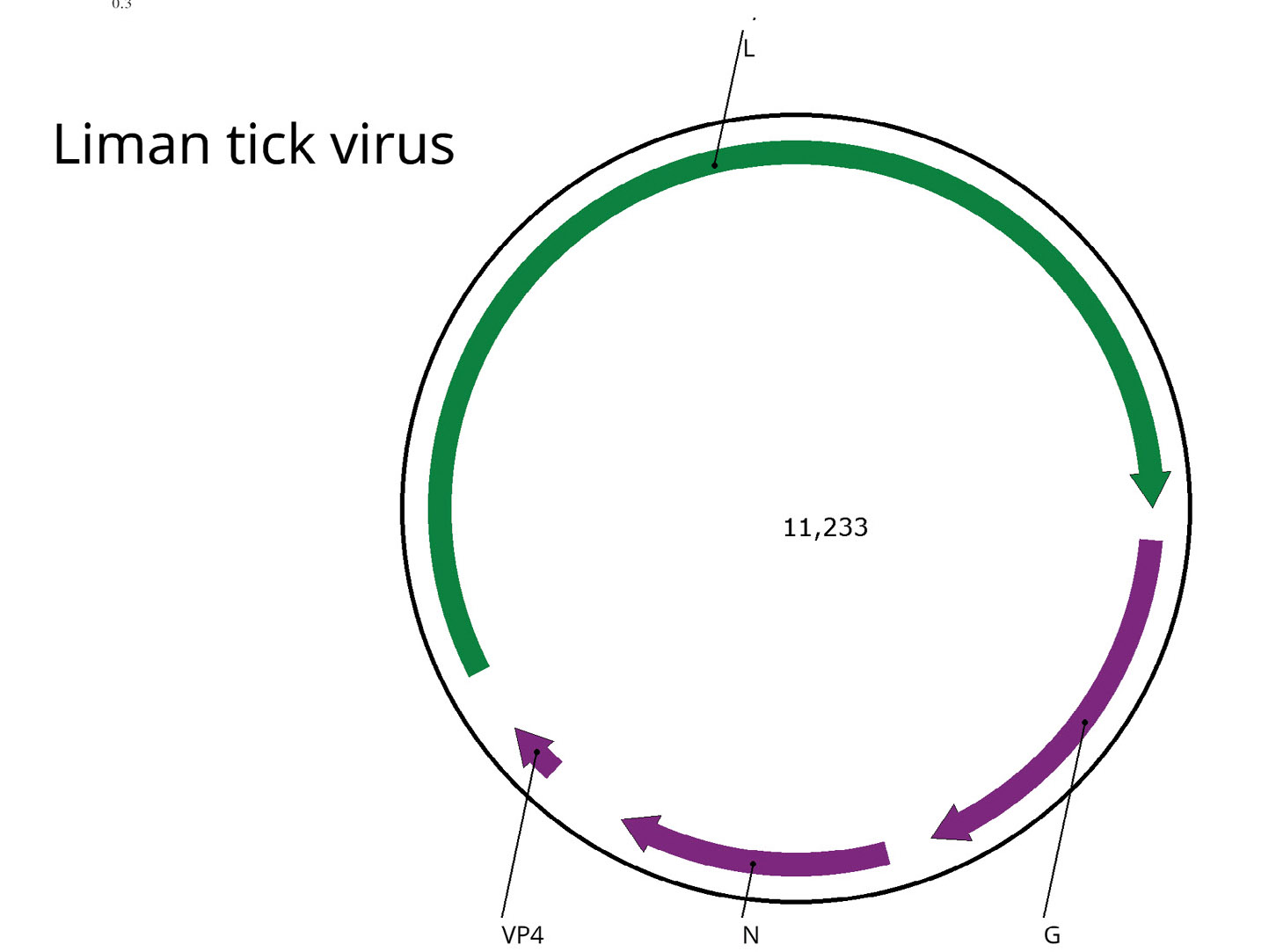
A detailed analysis of the genome assembly and its comparison with the reference genome allowed us to establish that in our case there are direct repeats at the ends of the contig, which indicates the circular nature of the LMTV genome. This has been repeatedly described for representatives of the Chuviridae family [17] and shown earlier for LMTV [30]. Thus, we were able to assemble the complete genome of this virus. The genome map is presented in Figure (b). The genome of the virus has been deposited in the international GenBank database under the number PQ613839.
Figure. Genomic structure and phylogenetic relationships of the Liman tick virus. a ‒ Midpoint-rooted phylogenetic tree of the family Chuviridae. Maximum-likelihood phylogenetic tree was constructed using the amino acid sequences of the RdRp (1000 bootstrap replicates; nodes with ≥ 85% bootstrap support are indicated). The scale bar represents the number of amino acid substitutions per site. The Liman tick viruses clade is marked in yellow. Liman tick virus detected in the current work marked in red; b ‒ Scheme of the Liman tick virus genome. ORFs are shown in purple. The RdRp-encoding ORF is marked in green.
Рисунок. Структура генома и филогенетические взаимоотношения вируса Liman tick. а ‒ укорененное в среднюю точку филогенетическое дерево семейства Chuviridae. Дерево было построено методом максимального правдоподобия с использованием аминокислотных последовательностей РНК-зависимой РНК-полимеразы (1000 реплик bootstrap; указаны узлы с поддержкой bootstrap ≥ 85%). Шкала представляет количество аминокислотных замен на сайт. Клада Liman tick вирусов выделена желтым цветом. Liman tick вирус, обнаруженный в данной работе, выделен красным цветом; б ‒ схема генома Liman tick вируса. Открытые рамки считывания (ОРС) показаны фиолетовым цветом. ОРС, кодирующая вирусную полимеразу, отмечена зеленым цветом.
The number of LMTV reads in the sample was quite high, 2.42% relative to the total reads, which is an indirect indicator that the virus is actively replicating during persistence in HAE/CTVM8 cell culture.
Phylogenetic analysis based on RNA-dependent RNA polymerase sequences showed that the detected virus was a close relative and formed a monophyletic group with other LMTV isolates from ticks of the Hyalomma genus. The LMTV group itself formed a monophyletic group with isolates of Bole tick virus 3 (Mivirus boleense) and fell into the Mivirus genus.
Discussion
HAE/CTVM8 cell culture is routinely used for virological studies [8, 18–20]. Despite the fact that, according to some reports, this culture contains persistent viruses [10], their genomes have remained unknown until now, and the culture itself has not been investigated using modern approaches. Previously, we described the presence of three rhabdoviruses in IRE/CTVM19 cell culture [12].
In the present study, using a similar methodology, we detected the LMTV genome in HAE/CTVM8 cell culture, which was obtained in 1991, indicating the long-term presence of LMTV in this system. Moreover, since the assembled genome is complete and circular, its incorporation into the host genome is highly unlikely. The present study confirms the effectiveness of the metagenomic approach in characterizing newly obtained cell cultures.
The LMTV genome was first detected in H. anatolicum ticks collected in Liman district, Astrakhan region, Russian Federation, and published in GenBank in 2020 by Dr. Alkhovsky et al. (MN542376.1). Later, a new isolate of this virus was detected in H. rufipes ticks collected from camels in Garrisa County, Kenya. Moreover, antibodies to LMTV were detected in one of the camels studied and the virus itself was detected in blood [30]. These data indicate that this virus may be an arbovirus.
According to bioinformatic data, Chuviridae has an extremely interesting biology, as it includes viruses with circular single-stranded (–)RNA. At the same time, replication and virion structure of representatives of the Chuviridae family remain unexplored [17]. Thus, HAE/CTVM8 cell culture is a ready model for studying replication and biology of the Chuviridae family.
There are well described examples when the presence of one virus in cell culture has a significant effect on the reproduction of other viruses, both closely related and phylogenetically distant viruses [31–33]. It remains unclear how LMTV may interact with other viruses, but the presence of this virus in HAE/CTVM8 cell culture should certainly be taken into account when conducting further experiments.
Conclusion
The information obtained in this study is essential for the use of HAE/CTVM8 cell culture in research and isolation of new viruses. The HAE/CTVM8 cell line with persistent LMTV is a system that is sufficient for studying the reproduction of Chuviridae representatives.
About the authors
Alexander G. Litov
Chumakov Federal Scientific Center for Research and Development of Immune-and-Biological Products of RAS (Institute of Poliomyelitis); Institute for Translational Medicine and Biotechnology, Sechenov University
Author for correspondence.
Email: novosti-wxo@yandex.ru
ORCID iD: 0000-0002-6086-3655
PhD, leading researcher in Laboratory of Biology of Arboviruses
Russian Federation, 108819, Moscow; 119991, MoscowAlexey M. Shchetinin
Gamaleya Federal Research Centre for Epidemiology and Microbiology, Ministry of Health of the Russian Federation
Email: shchetinin.alexey@yandex.ru
ORCID iD: 0000-0003-1842-3899
researcher in Pathogenic Microorganisms Variability Laboratory
Russian Federation, 123098, MoscowIvan S. Kholodilov
Chumakov Federal Scientific Center for Research and Development of Immune-and-Biological Products of RAS (Institute of Poliomyelitis)
Email: ivan-kholodilov@bk.ru
ORCID iD: 0000-0002-3764-7081
MD, PhD, leading researcher in Laboratory of Biology of Arboviruses
Russian Federation, 108819, MoscowOxana A. Belova
Chumakov Federal Scientific Center for Research and Development of Immune-and-Biological Products of RAS (Institute of Poliomyelitis)
Email: mikasusha@bk.ru
ORCID iD: 0000-0002-9040-0774
PhD, leading researcher in Laboratory of Biology of Arboviruses
Russian Federation, 108819, MoscowАnna S. Kalyanova
Chumakov Federal Scientific Center for Research and Development of Immune-and-Biological Products of RAS (Institute of Poliomyelitis)
Email: annakalyanova@bk.ru
ORCID iD: 0009-0003-1154-3852
junior researcher in Laboratory of Biology of Arboviruses
Russian Federation, 108819, MoscowVladimir A. Gushchin
Gamaleya Federal Research Centre for Epidemiology and Microbiology, Ministry of Health of the Russian Federation; Federal State Autonomous Educational Institution of Higher Education I.M. Sechenov First Moscow State Medical University of the Ministry of Health of the Russian Federation (Sechenov University)
Email: wowaniada@gmail.com
ORCID iD: 0000-0002-9397-3762
Doctor of Biological Sciences, leading researcher and head of Pathogenic Microorganisms Variability Laboratory
Russian Federation, 123098, Moscow; 119991, MoscowGalina G. Karganova
Chumakov Federal Scientific Center for Research and Development of Immune-and-Biological Products of RAS (Institute of Poliomyelitis); Institute for Translational Medicine and Biotechnology, Sechenov University
Email: karganova@bk.ru
ORCID iD: 0000-0002-8901-6206
Doctor of Biological Sciences, professor, leading researcher and head of Laboratory of Biology of Arboviruses
Russian Federation, 108819, Moscow; 119991, MoscowReferences
- Marques A.R., Strle F., Wormser G.P. Comparison of Lyme disease in the United States and Europe. Emerg. Infect. Dis. 2021; 27(8): 2017–24. https://doi.org/10.3201/eid2708.204763
- Ruzek D., Avšič Županc T., Borde J., Chrdle A., Eyer L., Karganova G., et al. Tick-borne encephalitis in Europe and Russia: Review of pathogenesis, clinical features, therapy, and vaccines. Antiviral. Res. 2019; 164: 23–51. https://doi.org/10.1016/j.antiviral.2019.01.014
- Bell-Sakyi L., Zweygarth E., Blouin E.F., Gould E.A., Jongejan F. Tick cell lines: tools for tick and tick-borne disease research. Trends. Parasitol. 2007; 23(9): 450–7. https://doi.org/10.1016/j.pt.2007.07.009
- Mangia C., Vismarra A., Kramer L., Bell-Sakyi L., Porretta D., Otranto D., et al. Evaluation of the in vitro expression of ATP binding-cassette (ABC) proteins in an Ixodes ricinus cell line exposed to ivermectin. Parasit. Vectors. 2016; 9: 215. https://doi.org/10.1186/s13071-016-1497-2
- Kholodilov I.S., Litov A.G., Klimentov A.S., Belova O.A., Polienko A.E., Nikitin N.A., et al. Isolation and characterisation of Alongshan virus in Russia. Viruses. 2020; 12(4): 362. https://doi.org/10.3390/v12040362
- Palomar A.M., Premchand-Branker S., Alberdi P., Belova O.A., Moniuszko-Malinowska A., Kahl O., et al. Isolation of known and potentially pathogenic tick-borne microorganisms from European ixodid ticks using tick cell lines. Ticks Tick Borne Dis. 2019; 10(3): 628–38. https://doi.org/10.1016/j.ttbdis.2019.02.008
- Husin N.A., Khoo J.J., Zulkifli M.M.S., Bell-Sakyi L., AbuBakar S. Replication kinetics of Rickettsia raoultii in tick cell lines. Microorganisms. 2021; 9(7): 1370. https://doi.org/10.3390/microorganisms9071370
- Salata C., Moutailler S., Attoui H., Zweygarth E., Decker L., Bell-Sakyi L. How relevant are in vitro culture models for study of tick-pathogen interactions? Pathog. Glob. Health. 2021; 115(7-8): 437–55. https://doi.org/10.1080/20477724.2021.1944539
- Bell-Sakyi L. Continuous cell lines from the tick Hyalomma anatolicum anatolicum. J. Parasitol. 1991; 77(6): 1006–8.
- Alberdi M.P., Dalby M.J., Rodriguez-Andres J., Fazakerley J.K., Kohl A., Bell-Sakyi L. Detection and identification of putative bacterial endosymbionts and endogenous viruses in tick cell lines. Ticks Tick Borne. Dis. 2012; 3(3): 137–46. https://doi.org/10.1016/j.ttbdis.2012.05.002
- Attoui H., Stirling J.M., Munderloh U.G., Billoir F., Brookes S.M., Burroughs J.N., et al. Complete sequence characterization of the genome of the St Croix River virus, a new orbivirus isolated from cells of Ixodes scapularis. J. Gen. Virol. 2001; 82(Pt. 4): 795–804. https://doi.org/10.1099/0022-1317-82-4-795
- Litov A.G., Shchetinin A.M., Kholodilov I.S., Belova O.A., Gadzhikurbanov M.N., Ivannikova A.Y., et al. High-throughput sequencing reveals three rhabdoviruses persisting in the IRE/CTVM19 cell line. Viruses. 2024; 16(4): 576. https://doi.org/10.3390/v16040576
- Harvey E., Rose K., Eden J.S., Lo N., Abeyasuriya T., Shi M., et al. Extensive diversity of RNA viruses in Australian ticks. J. Virol. 2019; 93(3): e01358–18. https://doi.org/10.1128/JVI.01358-18
- Li C.X., Shi M., Tian J.H., Lin X.D., Kang Y.J., Chen L.J., et al. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife. 2015; 4: e05378. https://doi.org/10.7554/eLife.05378
- Ni X.B., Cui X.M., Liu J.Y., Ye R.Z., Wu Y.Q., Jiang J.F., et al. Metavirome of 31 tick species provides a compendium of 1,801 RNA virus genomes. Nat. Microbiol. 2023; 8(1): 162–73. https://doi.org/10.1038/s41564-022-01275-w
- Shi M., Lin X.D., Tian J.H., Chen L.J., Chen X., Li C.X., et al. Redefining the invertebrate RNA virosphere. Nature. 2016; 540(7634): 539–43. https://doi.org/10.1038/nature20167
- Kuhn J.H., Dheilly N.M., Junglen S., Paraskevopoulou S., Shi M., Di Paola N. ICTV virus taxonomy profile: Jingchuvirales 2023. J. Gen. Virol. 2023; 104(12): 001924. https://doi.org/10.1099/jgv.0.001924
- Kholodilov I.S., Belova O.A., Ivannikova A.Y., Gadzhikurbanov M.N., Makenov M.T., Yakovlev A.S., et al. Distribution and characterisation of tick-borne flavi-, flavi-like, and phenuiviruses in the Chelyabinsk region of Russia. Viruses. 2022; 14(12): 2699. https://doi.org/10.3390/v14122699
- Salata C., Monteil V., Karlberg H., Celestino M., Devignot S., Leijon M., et al. The DEVD motif of Crimean-Congo hemorrhagic fever virus nucleoprotein is essential for viral replication in tick cells. Emerg. Microbes Infect. 2018; 7(1): 190. https://doi.org/10.1038/s41426-018-0192-0
- Salvati M.V., Salaris C., Monteil V., Del Vecchio C., Palù G., Parolin C., et al. Virus-derived DNA forms mediate the persistent infection of tick cells by Hazara virus and Crimean-Congo hemorrhagic fever virus. J. Virol. 2021; 95(24): e0163821. https://doi.org/10.1128/JVI.01638-21
- Kholodilov I.S., Belova O.A., Morozkin E.S., Litov A.G., Ivannikova A.Y., Makenov M.T., et al. Geographical and tick-dependent distribution of flavi-like Alongshan and Yanggou tick viruses in Russia. Viruses. 2021; 13(3): 458. https://doi.org/10.3390/v13030458
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30(15): 2114–20. https://doi.org/10.1093/bioinformatics/btu170
- Lo C.C., Chain P.S. Rapid evaluation and quality control of next generation sequencing data with FaQCs. BMC Bioinformatics. 2014; 15(1): 366. https://doi.org/10.1186/s12859-014-0366-2
- Litov A.G., Semenyuk I.I., Belova O.A., Polienko A.E., Thinh N.V., Karganova G.G., et al. Extensive diversity of viruses in millipedes collected in the Dong Nai biosphere reserve (Vietnam). Viruses. 2024; 16(9): 1486. https://doi.org/10.3390/v16091486
- Okonechnikov K., Golosova O., Fursov M. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012; 28(8): 1166–7. https://doi.org/10.1093/bioinformatics/bts091
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013; 30(4): 772–80. https://doi.org/10.1093/molbev/mst010
- Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009; 25(15): 1972–3. https://doi.org/10.1093/bioinformatics/btp348
- Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015; 32(1): 268–74. https://doi.org/10.1093/molbev/msu300
- Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017; 14(6): 587–9. https://doi.org/10.1038/nmeth.4285
- Zhang Y., Hu B., Agwanda B., Fang Y., Wang J., Kuria S., et al. Viromes and surveys of RNA viruses in camel-derived ticks revealing transmission patterns of novel tick-borne viral pathogens in Kenya. Emerg. Microbes Infect. 2021; 10(1): 1975–87. https://doi.org/10.1080/22221751.2021.1986428
- Abrao E.P., da Fonseca B.A. Infection of mosquito cells (C6/36) by Dengue-2 virus interferes with subsequent infection by yellow fever virus. Vector. Borne Zoonotic Dis. 2016; 16(2): 124–30. https://doi.org/10.1089/vbz.2015.1804
- Kuwata R., Isawa H., Hoshino K., Sasaki T., Kobayashi M., Maeda K., et al. Analysis of mosquito-borne flavivirus superinfection in Culex tritaeniorhynchus (Diptera: Culicidae) cells persistently infected with Culex flavivirus (Flaviviridae). J. Med. Entomol. 2015; 52(2): 222–9. https://doi.org/10.1093/jme/tju059
- Patterson E.I., Kautz T.F., Contreras-Gutierrez M.A., Guzman H., Tesh R.B., Hughes G.L., et al. Negeviruses reduce replication of alphaviruses during coinfection. J. Virol. 2021; 95(14): e0043321. https://doi.org/10.1128/JVI.00433-21
Supplementary files