Analysis of the association of influenza clinical course with single nucleotide polymorphisms in genes affecting the interferon-λ3 production
- Authors: Nikolaeva L.I.1, Stuchinskaya M.D.1, Telepenina K.P.1, Shevchenko N.G.1, Kuprianov V.V.1, Krasnoslobodtsev K.G.1, Mukasheva E.A.1, Trushakova S.V.1, Khlopova I.N.1, Kruzhkova I.S.1, Kisteneva L.B.1, Kolobukhina L.V.1, Burtseva E.I.1
-
Affiliations:
- N.F. Gamaleya National Research Center of Epidemiology and Microbiology of the Ministry of Health of Russia
- Issue: Vol 70, No 1 (2025)
- Pages: 25-34
- Section: ORIGINAL RESEARCHES
- URL: https://virusjour.crie.ru/jour/article/view/16691
- DOI: https://doi.org/10.36233/0507-4088-271
- EDN: https://elibrary.ru/hnrscv
- ID: 16691
Cite item
Abstract
Introduction. Predisposition to different courses of the infectious process is largely associated with the polymorphisms in human genome, especially in genes encoding proteins of the immune system. In the early stages of influenza infection such components of innate immunity as interferons I (α/β) and III (λ) type play a significant role in limiting virus replication.
The aim of the work was to investigate associations of single nucleotide polymorphism in IFNL3 (rs8099917 T/G) and IFNL4 (rs12979860 C/T) genes with different course of influenza, and identify genetic markers of influenza complicated by community-acquired pneumonia. The genes noted above affect the production of interferon-λ3, which is involved in restriction of the viral replication.
Materials and methods. Samples from 456 patients with mild (n = 150), moderate (n = 173), and severe (n = 133) influenza were studied. The viral RNA was detected by reverse transcription and polymerase chain reaction (RT-PCR). Polymorphisms in IFNL3 (rs8099917 T/G) and IFNL4 (rs12979860 C/T) genes was detected by PCR. Statistical analysis was performed using SNPStats software.
Results. Patients with the C/T or T/T genotype of IFNL4 gene (rs12979860 C/T) were more likely to have pneumonia than those with the C/C genotype (OR 2.47 (1.31–4.63); p = 0.0044; q = 0.0059). The presence of one T allele increased the risk of developing pneumonia (OR 2.02 (1.05–4.02); p = 0.006; q = 0.008). In the presence of the T/T genotype, the risk increased more than twofold: OR 2.14 (1.31–3.48). Analysis of the SNP of IFNL3 gene (rs8099917 T/G) revealed a weak association of the G allele with pneumonia (OR 1.86 (1.04–3.31); p = 0.03; q = 0.045).
Conclusion. Genetic markers of increased risk of community-acquired pneumonia in influenza include the presence of the T allele in IFNL4 gene (rs12979860 C/T) and, to a lesser extent, the G allele in IFNL3 gene (rs8099917 T/G). Patients carrying these alleles have an increased risk of developing pneumonia, especially in old age.
Full Text
Introduction
Influenza A and B viruses cause annual epidemic outbreaks in humans. Influenza A viruses are also responsible for epizootics among birds and certain mammalian species. Annually, about 1 billion cases of influenza in humans are registered worldwide, of which 3‒5 million have a severe course; the number of fatalities varies from 290 to 650 thousand cases, depending on the season1 [1]. In the Russian Federation, influenza and other respiratory viral infections are registered annually in an average of 30 million people, and the annual economic loss is about 40 billion rubles [2].
Influenza is most often complicated by the development of out-of-hospital pneumonia, which is detected in a significant proportion of hospitalized patients and can be fatal [3]. After the end of the influenza epidemic within 2‒3 months, additional (delayed) mortality from this infection is registered in patients from risk groups with a history of heart, lung and other diseases. Experts have calculated that mortality after influenza can increase in persons with chronic cardiovascular pathology by 52 times, and with chronic lung diseases – 120 times [4].
The last influenza pandemic declared by the World Health Organization in 2009 was caused by the A(H1N1)pdm09 strain, which had genes from swine, avian and seasonal human influenza in its genome structure. The A(H1N1)pdm09 pandemic affected all age groups and had more severe consequences than previous ones. Currently, avian influenza virus type A, which has hemagglutin variants H5 and H7, poses an obvious danger [5]. Cases of human infection with these influenza viruses after contact with sick birds have been registered, which ended lethally in 30‒50% of patients. The causes of lethal outcomes of infection with avian influenza A(H5N1) virus are considered to be the tropism of the virus to receptors of tissues of different organs, its more intensive replication and hyperinflammatory immune response of infected people [5, 6].
The reactions of the human immune system to many pathogens and influenza antigens, in particular, depend on the variability (polymorphism) of the genome, which determines a certain predisposition to different course of the infectious process. Type I and III interferons (IFNs) play a significant role in limiting influenza virus replication in the early stages of infection. As components of innate immunity, they trigger the production of cytokines, chemokines, stimulate the attraction of neutrophils, monocytes, NK-cells to the focus of inflammation and induce the biosynthesis of intracellular antiviral proteins before the formation of an adaptive immune response.
Type III IFNs were identified in 2003 by analyzing new proteins discovered through the Human Genome Project and classified as interleukin (IL) 28A, IL-28B, and IL-29 [7]. Subsequently, in 2012, these cytokines were renamed IFN-λ1 (IL-29), IFN-λ2 (IL-28A), and IFN-λ3 (IL-28B) on the recommendation of the nomenclature committee at HUGO (Human Genome Organization). It was found that type III IFNs are produced slightly earlier than type I IFNs and provide protection of the mucosal barrier of the airway epithelium [8, 9]. Recombinant type III IFNs can effectively limit influenza infection in laboratory mice [10, 11]. The possibility of creating a drug based on a PEGylated form of type III IFN for therapy of viral respiratory disease COVID-19 has been demonstrated [12].
Single nucleotide polymorphisms (SNPs) affecting the expression of this cytokine have been identified near the gene encoding IFN-λ3 [13]. The most significant SNPs for a number of viral infections are localized upstream of the IFNL3 gene (rs8099917 T/G), in the intron of the IFNL4 gene (rs12979860 C/T) and in the exon of the IFNL4 gene (rs368234815 ΔG/TT) [7]. The serum IFN-λ3 content is influenced by SNPs in IFNL3 (rs8099917 T/G) and IFNL4 (rs12979860 C/T) genes. Taking into account the above-mentioned and the fact that the contribution of SNPs of the above genes to the development of severe course of influenza in our country residents is practically not studied, we chose to analyze potentially possible associations of IFNL3 (rs8099917 T/G) and IFNL4 (rs12979860 C/T) gene loci with clinical variants of influenza infection course in the current study.
The main aim of the study was to identify markers of severe influenza course by variant loci of IFNL3 (rs8099917 T/G) and IFNL4 (rs12979860 C/T) genes.
Materials and methods
Patients and groups. Biological material (blood and nasopharyngeal swabs) was collected between October 2020 and May 2024 from adult patients. All patients had a virologically confirmed diagnosis of influenza. The study was conducted with voluntary informed consent of the patients. The study protocol was approved by the ethical committee of the Infectious Disease Clinical Hospital No. 1 (protocol No. 11/A of 16.10.2020 and No. 8 of 28.12.2022). Group 1 included 150 patients who had mild influenza. The age range was 20‒84 years, mean age was 44.6 years, male to female ratio: 46.7% to 53.3%. Group 2 included 173 patients hospitalized with moderate influenza without out-of-hospital pneumonia. The age range was 18‒89 years, mean age 41.4 years, male to female ratio: 46.8% to 53.2%. Group 3 included 133 patients hospitalized with severe influenza infection and out-of-hospital pneumonia. The age range was 18‒94 years of age, the mean age was 53 years of age, the ratio of men to women: 57.1% to 42.9%. The analyzed groups of patients were comparable in terms of gender composition and age range (for all groups p > 0.05). The total number of patients was 456 (226 males and 230 females), which allowed us to obtain statistical data with a significance level of 0.05 and a confidence interval of 4.59 [14].
Influenza virus genome identification and typing in nasopharyngeal swabs was performed by a method combining reverse transcription and polymerase chain reaction (RT-PCR) using AmpliSens Influenza viruses A/B, AmpliSens Influenza viruses A/H1-swine-FL, AmpliSens Influenza viruses A-type FL test systems according to the manufacturer’s recommendations (Central Research Institute for Influenza Virus Research, Russia).
To analyze allelic variants of polymorphic regions of IFNL3 (rs8099917 T/G) and IFNL4 (rs12979860 C/T) genes, DNA was isolated from venous blood cells using the Proba-Rapid-Genetics reagent kit (DNA-Technology, Russia). Determination of genotypes in polymorphic regions of IFNL3 (rs8099917 T/G) and IFNL4 (rs12979860 C/T) genes was performed by PCR using the Immunogenetics IL28B reagent kit (DNA-Technology, Russia).
Statistical analysis was carried out with the Statistica v. 10 software package (StatSoft, USA). To assess the reliability of differences of analyzed parameters in patient groups, the χ2 criterion and Fisher’s exact method were used (if one of the parameters was less than 10). SNPStats program (https://www.snpstats.net/) was used to analyze the association of allelic variants of polymorphic regions of IFNL3 (rs8099917 T/G) and IFNL4 (rs12979860 C/T) genes in different groups of patients. Groups were compared according to 5 possible inheritance patterns: codominant, dominant, recessive, overdominant and log-additive. The correction q-value for multiple comparisons (to estimate the probability of rejection of the null hypothesis) was calculated using the software at https://www.sdmproject.com/utilities/?show=FDR. To assess the significance of differences, 95% confidence interval (95% CI), odds ratio (OR) and Akaike information criterion (AIC) were calculated. The differences were considered significant at p < 0.05.
Results
The first stage of the study involved the comparison of genetic indices for the analyzed genes in groups of patients differing in the clinical course of influenza infection. For this purpose, the genotyping data of group 1 (mild influenza) were compared with the corresponding indicators of group 2 (moderate influenza, Table 1) and with the indicators of group 3 (severe influenza, Table 2).
Table 1. Results of genotype analysis in polymorphic loci of IFNL3 (rs8099917 T/G) and IFNL4 (rs12979860 C/T) genes in patients with mild and moderate influenza
Таблица 1. Результаты анализа генотипов в полиморфных локусах генов IFNL3 (rs8099917 T/G) и IFNL4 (rs12979860 C/T) у пациентов с легким и среднетяжелым течением гриппа
Analysis model Модель aнализа | Genotypes Генотипы | 1st group (n = 150), abs. (%) 1-я группа (n = 150), абс. (%) | 2nd group (n = 173), abs. (%) 2-я группа (n = 173), абс. (%) | OR (95% CI) ОШ (95% ДИ) | p-value Значение р | AIC ИКА |
Gene / Ген IFNL4 | ||||||
Codominant Кодоминантная | С/С C/T T/T | 77 (51.3) 58 (38.7) 15 (10.0) | 94 (54.3) 63 (36.4) 16 (9.2) | 1.00 1.12 (0.70–1.79) 1.14 (0.53–2.46) | 0.86 | 451.8 |
Dominant Доминантная | С/С С/Т – T/T | 77 (51.3) 73 (48.7) | 94 (54.3) 79 (45.7) | 1.00 1.13 (0.73–1.75) | 0.59 | 449.8 |
Recessive Рецессивная | C/C – C/T T/T | 135 (90.0) 15 (10.0) | 157 (90.8) 16 (9.2) | 1.00 1.09 (0.52–2.29) | 0.83 | 450.1 |
Overdominant Сверхдоминантная | С/С – Т/Т C/T | 92 (61.3) 58 (38.7) | 110 (63.6) 63 (36.4) | 1.00 1.10 (0.70–1.73) | 0.68 | 450 |
Log-additive Лог-аддитивная | – | – | – | 1.09 (0.78–1.52) | 0.61 | 449.9 |
Gene / Ген IFNL3 | ||||||
Codominant Кодоминантная | T/T T/G G/G | 108 (72.0) 39 (26.0) 3 (2.0) | 123 (71.1) 46 (26.6) 4 (2.3) | 1.00 0.97 (0.59–1.59) 0.85 (0.19–3.90) | 0.97 | 452.1 |
Dominant Доминантная | T/T T/G – G/G | 108 (72.0) 42 (28.0) | 123 (71.1) 50 (28.9) | 1.00 0.96 (0.59–1.55) | 0.86 | 450.1 |
Recessive Рецессивная | T/T – T/G G/G | 147 (98.0) 3 (2.0) | 169 (97.7) 4 (2.3) | 1.00 0.86 (0.19–3.92 | 0.85 | 450.1 |
Overdominant Сверхдоминантная | T/T – G/G T/G | 111 (74.0) 39 (26.0) | 127 (73.4) 46 (26.6) | 1.00 0.97 (0.59–1.59) | 0.9 | 450.1 |
Log-additive Лог-аддитивная | – | – | – | 0.95 (0.62–1.47) | 0.83 | 450.1 |
Table 2. Results of genotype analysis in polymorphic loci of IFNL3 (rs8099917 T/G) and IFNL4 (rs12979860 C/T) genes in patients with mild and severe influenza
Таблица 2. Результаты анализа генотипов в полиморфных локусах генов IFNL3 (rs8099917 T/G) и IFNL4 (rs12979860 C/T) у пациентов с легким и тяжелым течением гриппа
Analysis model Модель aнализа | Genotyp Генотипы | 1st group (n = 150), abs. (%) 1-я группа (n = 150), абс. (%) | 3rd group (n = 133), abs. (%) 3-я группа (n = 133), абс. (%) | OR (95% CI) ОШ (95% ДИ) | p-value Значение | AIC ИКА |
Gene / Ген IFNL4 | ||||||
Codominant Кодоминантная | С/С C/T T/T | 77 (51.3) 58 (38.7) 15 (10.0) | 61 (45.9) 53 (39.9) 19 (14.3) | 1.00 1.15 (0.70–1.90) 1.60 (0.75–3.40) | 0.46 | 395.8 |
Dominant Доминантная | С/С С/Т – T/T | 77 (51.3) 73 (48.7) | 61 (45.9) 72 (54.1) | 1.00 1.25 (0.78–1.99) | 0.36 | 394.5 |
Recessive Рецессивная | C/C – C/T T/T | 135 (90.0) 15 (10.0) | 114 (85.7) 19 (14.3) | 1.00 1.50 (0.73–3.09) | 0.27 | 394.1 |
Overdominant Сверхдоминантная | С/С – Т/Т C/T | 92 (61.3) 58 (38.7) | 80 (60.1) 53 (39.9) | 1.00 1.05 (0.65–1.69) | 0.84 | 395.3 |
Log-additive Лог-аддитивная | – | – | – | 1.23 (0.87–1.73) | 0.23 | 393.9 |
Gene / Ген IFNL3 | ||||||
Codominant Кодоминантная | T/T T/G G/G | 108 (72.0) 39 (26.0) 3 (2.0) | 85 (63.9) 42 (31.6) 6 (4.5) | 1.00 1.37 (0.81–2.30) 2.54 (0.62–10.46) | 0.24 | 393.4 |
Dominant Доминантная | T/T T/G – G/G | 108 (72.0) 42 (28.0) | 85 (63.9) 48 (36.1) | 1.00 1.45 (0.88–2.40) | 0.14 | 393.2 |
Recessive Рецессивная | T/T – T/G G/G | 147 (98.0) 3 (2.0) | 127 (95.5) 6 (4.5) | 1.00 2.31 (0.57–9.45) | 0.23 | 393.8 |
Overdominant Сверхдоминантная | T/T – G/G T/G | 111 (74.0) 39 (26.0) | 91 (68.4) 42 (31.6) | 1.00 1.31 (0.78–2.20) | 0.3 | 394.2 |
Log-additive Лог-аддитивная | – | – | – | 1.44 (0.93–2.23) | 0.099 | 393.6 |
As shown by the data in Table 1, there were no significant differences in the two groups.
As shown by the data of Tables 1 and 2, no significant differences in the frequency of occurrence of individual genotypes were found in three compared groups differing in the clinical course of influenza infection. However, there was a decrease in the proportion of CC genotype (rs12979860) in the IFNL4 gene and TT genotype (rs8099917) in the IFNL3 gene from group 1 to group 3.
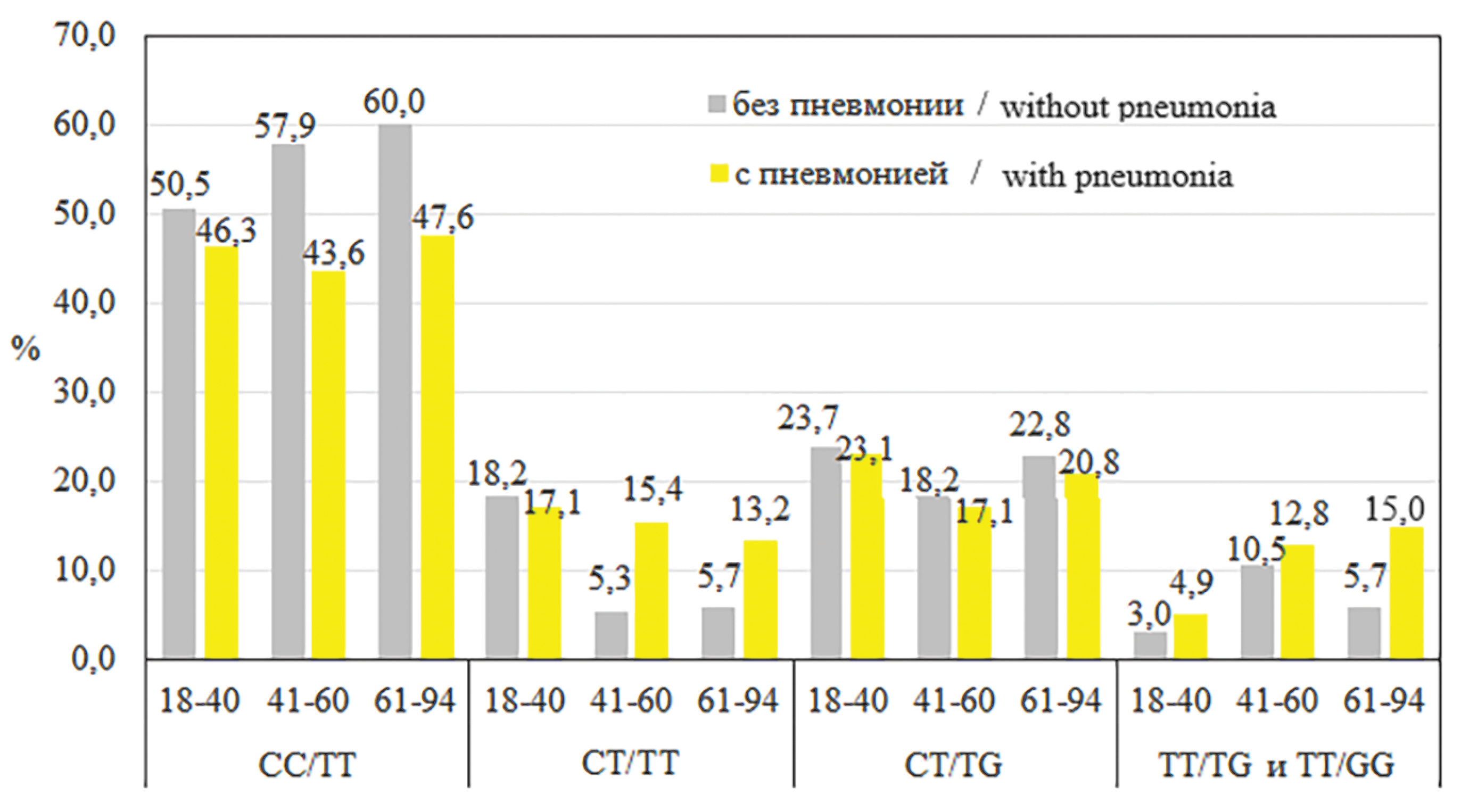
At the next stage of the study, another criterion was chosen: “with pneumonia” or “without pneumonia”, and genotypes of groups with moderate and severe course were compared. But beforehand, the influence of age on the development of pneumonia was evaluated. For this purpose, age subgroups were identified: 1) a subgroup consisting of patients 18‒40 years of age (n = 140), 2) 41‒60 years of age (n = 78), 3) 61‒94 years of age (n = 88). The Figure below shows the distribution of genotypes in the analyzed gene loci for all age subgroups of patients. It is visually noticeable that the CC/TT genotype is more frequently detected in individuals without pneumonia in all age subgroups. The emergence of the T allele changed the ratio of cases with and without pneumonia, the ST/TT genotype became more often detected in persons with pneumonia over 40 years of age. With the ST/TG genotype, the frequency of cases with and without pneumonia was very close. The frequencies of pneumonia detection at TT/TG, and TT/GG genotypes were close in the compared age subgroups.
Figure. Distribution of genotypes by loci of the IFNL3 (rs8099917 T/G) and IFNL4 (rs12979860 C/T) genes, taking into account age subgroups.
The percentages are calculated for the entire group. The TT/TT genotype is not included due to the small number.
Рисунок. Распределение генотипов по локусам генов IFNL3 (rs8099917 T/G) и IFNL4 (rs12979860 C/T) с учетом возрастных подгрупп.
Проценты рассчитаны на всю группу. Генотип ТТ/ТТ не включен из-за малочисленности.
The proportion of patients with pneumonia in the 18‒40 age subgroup was 29.3% (n = 41), in the 41‒60 age subgroup was 51.3% (n = 40), and in the 61‒94 age subgroup was 60.2% (n = 53), The incidence of pneumonia in the 18‒40 age subgroup was significantly lower than in the 41‒60 age subgroup (p = 0.0021) and in the 61‒94 age subgroup (p < 0.0001). Given this difference in pneumonia detection, we further analyzed the SNPs of IFNL3 (rs8099917 T/G) and IFNL4 (rs12979860 C/T) genes in the subgroup of patients up to and including 40 years of age (n = 140, pneumonia in 41), and in the combined subgroups of patients over 40 years of age (n = 166, pneumonia in 93). In the 18‒40 years of age subgroup, no significant differences were found when analyzing the SNPs of IFNL3 (rs8099917 T/G) and IFNL4 (rs12979860 C/T) genes in patients with pneumonia relative to participants without pneumonia (data not shown). The results of a similar analysis of patients aged 41 years and older for the pooled subgroups are presented in Table 3.
Table 3. Results of genotype analysis in polymorphic loci of IFNL3 (rs8099917 T/G) and IFNL4 (rs12979860 C/T) genes in patients over 41 years of age
Таблица 3. Результаты анализа генотипов в полиморфных локусах генов IFNL3 (rs8099917 T/G) и IFNL4 (rs12979860 C/T) у пациентов старше 41 года
Analysis model Модель aнализа | Genotyp Генотипы | Patients without pneumonia (n = 73), abs. (%) Пациенты без пневмонии (n = 73), абс. (%) | Patients with pneumonia (n = 93), abs. (%) Пациенты с пневмонией (n = 93), абс. (%) | OR (95% CI) ОШ (95% ДИ) | p-value (q) Значение р (q) | AIC ИКА |
Gene / Ген IFNL4 | ||||||
Codominant Кодоминантная | С/С C/T T/T | 46 (63.0) 23 (31.5) 4 (5.5) | 38 (40.9) 39 (41.9) 16 (17.2) | 1.00 2.02 (1.05–4.02) 4.84 (1.49–15.71) | 0.006 (0.008) | 223.5 |
Dominant Доминантная | С/С С/Т – T/T | 46 (63.0) 27 (37.0) | 38 (40.9) 55 (59.1) | 1.00 2.47 (1.31–4.63) | 0.0044 (0.0059) | 223.6 |
Recessive Рецессивная | C/C – C/T T/T | 69 (94.5) 4 (5.5) | 77 (82.8) 16 (17.2) | 1.00 3.58 (1.14–11.24) | 0.017 (0.0227) | 226 |
Overdominant Сверхдоминантная | С/С – Т/Т C/T | 50 (68.5) 23 (31.5) | 54 (58.1) 39 (41.9) | 1.00 1.57 (0.83–2.99) | 0.17 | 229.8 |
Log-additive Лог-аддитивная | – | – | – | 2.14 (1.31–3.48) | 0.0014 (0.0019) | 221.5 |
Gene / Ген IFNL3 | ||||||
Codominant Кодоминантная | T/T T/G G/G | 55 (75.3) 17 (23.3) 1 (1.4) | 58 (62.4) 28 (30.1) 7 (5.5) | 1.00 1.56 (0.77–3.17) | 0.066 | 228.3 |
Dominant Доминантная | T/T T/G – G/G | 55 (75.3) 18 (24.7) | 58 (62.4) 35 (37.6) | 1.00 1.84 (0.94–3.63) | 0.073 | 228.5 |
Recessive Рецессивная | T/T – T/G G/G | 72 (98.6) 1 (1.4) | 86 (92.5) 7 (7.5) | 1.00 5.86 (0.70–48.76 | 0.049 (0.065) | 227.8 |
Overdominant Сверхдоминантная | T/T – G/G T/G | 56 (76.7) 17 (23.3) | 65 (69.9) 28 (30.1) | 1.00 1.42 (0.70–2.86) | 0.32 | 230.0 |
Log-additive Лог-аддитивная | – | – | – | 1.00 1.86 (1.04–3.31) | 0.03 (0.045) | 227.0 |
The data in Table 3 show that patients with the C/T or T/T genotype at the polymorphic locus (rs12979860 C/T) of the IFNL4 gene were significantly more likely to develop pneumonia than those with genotype C/C (OR 2.47 (1.31‒4.63); p = 0.0044; q = 0.0059). Changes in the risk of pneumonia occurred with the occurrence of a single unfavorable T allele (OR 2.02 (1.05‒4.02); p = 0.006; q = 0.008). The log-additive model revealed a significant reliable association (p = 0.0014; q = 0.0019) and showed that two unfavorable alleles increased the risk of pneumonia more than 2-fold: OR 2.14 (1.31‒3.48). Allelic variants of the IFNL3 gene variant locus (rs8099917 T/G) showed a weak association of the G allele with pneumonia (p = 0.03; q = 0.045) in a log-additive model.
It is known that infection with influenza A(H1N1)pdm09 virus more often leads to severe course of influenza and development of pneumonia. In this regard, different strains of influenza virus were found in the analyzed groups and subgroups. Slight differences were observed in the group with moderate and severe course: influenza A(H3N2) virus was detected more often in the group with moderate course (49.1% vs. 40.6%), influenza A(H1N1)pdm09 virus was detected more often in patients with severe course (39.1% vs. 28.9%), but nowhere the level of statistically significant reliability was reached (p > 0.05). Influenza B/Victoria virus and one case of B/Yamagata (in a patient with pneumonia) were detected with similar frequency (22.0% vs. 21.8%). In patients older than 41 years of age, the incidence of these three virus strains also did not differ significantly (p = 0.0922 for A(H3N2); p = 0.1295 for A(H1N1)pdm09; p = 0.9771 for influenza B virus). Thus, virus strains could not significantly influence the development of pneumonia, which is important for the compared subgroups of patients older than 41 years. It has been established that the frequent development of pneumonia in A(H1N1)pdm09 virus infection is characteristic of the virus variant with mutations leading to the substitution of an asparagic acid residue for a tyrosine or asparagine residue in the receptor-binding site of hemagglutinin at position 222 [15]. However, this mutant variant of the A(H1N1)pdm09 virus has not been widely prevalent.
Discussion
Earlier, in 2017, domestic researchers A.A. Kurdin et al. analyzing the same genes and polymorphisms in 100 influenza patients with a moderate to severe course and 115 patients with a mild course of infection showed that these groups of patients differed significantly in the frequency of SS (2.5% vs. 79.1%), ST (95.0% vs. 21.7%) and TT (6.5% vs. 0%) genotype detection [16]. Our study included 456 patients, but no similar differences were obtained (Table 1). A.A. Kurdin et al. did not describe the method of genetic polymorphism detection and the characterization of primers or kits. In 2019, Iranian scientists M. Keshavarz et al., studying the polymorphism of inflammatory cytokine genes and IFNL3 gene (rs8099917 T/G) in 80 cases of severe influenza and 96 cases of influenza-like illnesses in the Iranian population, showed that the frequency of T and G alleles in the compared groups had no significant differences [17]. In our study, the number of participants was significantly larger, the population of our country has different genetic characteristics. Though the main point to take into account is that M. Keshavarz et al. made an unsuccessful choice of the comparison group. It was not contrasted to participants with severe influenza (it would have been optimal to use a group with mild influenza or people who rarely get influenza). In the study by M. Keshavarz et al., the comparison group was formed from patients with influenza-like illnesses.
According to the data of the present study, severe course of influenza infection complicated by community-acquired pneumonia was significantly less frequently observed in patients aged 18‒40 years of age than in patients of 41 years of age and older. In persons older than 60 years of age, out-of-hospital pneumonia was most frequently observed. Analysis of groups of patients over 41 years of age with moderate and severe course of infection showed that the association with the development of pneumonia was observed for allelic variants of the IFNL4 gene (rs12979860 C/T) and significantly weaker for the variant locus of the IFNL3 gene (rs8099917 T/G). Patients with genotype C/C (IFNL4, rs12979860 C/T) were significantly less likely to develop community-acquired pneumonia than carriers of other genotypes. Patients with the C/T genotype were significantly more likely to have pneumonia, i.e. the appearance of the T allele increased the risk of pneumonia. This pattern did not apply to younger participants (18‒40 years of age). It is known that at a young age the innate and adaptive immune response functions optimally [18]. Obviously, therefore, the risk of pneumonia development was reduced in the group of participants from 18 to 40 years of age. Age-matched patients have been shown to have a decreased adaptive immune response with preservation of the innate link [18]. Therefore, it is likely that we were able to detect the association of SNPs of IFNL4 (rs12979860 C/T) and IFNL3 (rs8099917 T/G) genes, influencing IFN-λ3 expression, with the development of pneumonia in influenza infection in patients older than 41 years of age. According to the data of the presented study, genetic markers of increased risk of developing out-of-hospital pneumonia in influenza are the presence of T allele in the variable locus of the IFNL4 gene (rs12979860 C/T) and, to a lesser extent, G allele in the polymorphic locus of the IFNL3 gene (rs8099917 T/G).
Conclusion
Influenza can be complicated by the development of community-acquired pneumonia, which is the most common cause of death in this infection. In this regard, the present study analyzed a number of factors influencing the development of pneumonia in influenza infection. One such factor is age. Persons over 40 years of age and especially over 60 years of age are most susceptible to influenza complicated by pneumonia, which once again emphasizes the need for annual influenza vaccination of this group of persons. The main aim of the presented study was to investigate the role of polymorphisms in innate immunity genes IFNL3 (rs8099917 T/G) and IFNL4 (rs12979860 C/T) affecting IFN-λ3 production in the development of pneumonia in influenza infection. It was found that of the two genes analyzed, the SNPs in the IFNL4 (rs12979860 C/T) gene were the most significant. It was shown that patients older than 41 years of age with genotype C/T or T/T at the polymorphic locus of the IFNL4 gene (rs12979860 C/T) were significantly more likely to develop pneumonia than those with genotype C/C (OR 2.47 (1.31‒4.63); p = 0.0044; q = 0.0059). The occurrence of one unfavorable T allele increased the risk of pneumonia (OR 2.02 (1.05‒4.02); p = 0.006; q = 0.008). Two unfavorable alleles increased the risk more than two-fold: OR 2.14 (1.31‒3.48); p = 0.0014; q = 0.0019. Allelic variants of the variable locus of the IFNL3 gene (rs8099917 T/G) showed a weaker association of the G allele with pneumonia (OR 1.86 (1.04‒3.31); p = 0.03; q = 0.045). Thus, genetic markers which have increased risk of developing out-of-hospital pneumonia in influenza are the presence of the T allele in the IFNL4 gene variable locus (rs12979860 C/T) and, to a lesser extent, the G allele in the IFNL3 gene polymorphic locus (rs8099917 T/G). This study is exploratory in nature and is aimed at developing a database of genetic markers for personalized predictive and preventive medicine.
1 World Health Organization. WHO launches new global influenza strategy (https://www.who.int/news/item/11-03-2019-who-launches-new-global-influenza-strategy).
About the authors
Lyudmila I. Nikolaeva
N.F. Gamaleya National Research Center of Epidemiology and Microbiology of the Ministry of Health of Russia
Author for correspondence.
Email: l.i.nikolaeva@mail.ru
ORCID iD: 0000-0002-1323-5568
D. Sci. in Biol., Leading Researcher of the Laboratory of Gene Engineering Products
Russian Federation, 123098, MoscowMaya D. Stuchinskaya
N.F. Gamaleya National Research Center of Epidemiology and Microbiology of the Ministry of Health of Russia
Email: mayastay@mail.ru
ORCID iD: 0000-0001-8544-7482
Junior Researcher of the Laboratory of Gene Engineering Products
Russian Federation, 123098, MoscowKristina P. Telepenina
N.F. Gamaleya National Research Center of Epidemiology and Microbiology of the Ministry of Health of Russia
Email: telepenina_kristina@mail.ru
ORCID iD: 0009-0003-3380-5104
Researcher Assistant of the Laboratory of Gene Engineering Products
Russian Federation, 123098, MoscowNadezhda G. Shevchenko
N.F. Gamaleya National Research Center of Epidemiology and Microbiology of the Ministry of Health of Russia
Email: dr.nadya@inbox.ru
ORCID iD: 0000-0002-2486-4554
Junior Researcher of the Laboratory of Gene Engineering Products
Russian Federation, 123098, MoscowVictor V. Kuprianov
N.F. Gamaleya National Research Center of Epidemiology and Microbiology of the Ministry of Health of Russia
Email: vkoop@mail.ru
ORCID iD: 0000-0002-8602-1974
C. Sci. in Biol., Senior Researcher of the Laboratory of Gene Engineering Products
Russian Federation, 123098, MoscowKirill G. Krasnoslobodtsev
N.F. Gamaleya National Research Center of Epidemiology and Microbiology of the Ministry of Health of Russia
Email: kkg_87@mail.ru
ORCID iD: 0000-0003-1745-9128
Researcher of the Laboratory of Influenza Etiology and Epidemiology
Russian Federation, 123098, MoscowEvgenya A. Mukasheva
N.F. Gamaleya National Research Center of Epidemiology and Microbiology of the Ministry of Health of Russia
Email: mukasheva_evgeniya@mail.ru
ORCID iD: 0000-0002-5688-5309
Researcher of the Laboratory of Influenza Etiology and Epidemiology
Russian Federation, 123098, MoscowSvetlana V. Trushakova
N.F. Gamaleya National Research Center of Epidemiology and Microbiology of the Ministry of Health of Russia
Email: s.trushakova@gmail.com
ORCID iD: 0000-0002-9610-3041
C. Sci. in Biol., Senior Researcher of the Laboratory Influenza Etiology and Epidemiology
Russian Federation, 123098, MoscowIrina N. Khlopova
N.F. Gamaleya National Research Center of Epidemiology and Microbiology of the Ministry of Health of Russia
Email: khlopova.ira@yandex.ru
ORCID iD: 0000-0002-7419-590X
C. Sci. in Med., Leading Researcher of the Laboratory of Chronic Viral Infections
Russian Federation, 123098 MoscowIrina S. Kruzhkova
N.F. Gamaleya National Research Center of Epidemiology and Microbiology of the Ministry of Health of Russia
Email: irina-kru@yandex.ru
ORCID iD: 0000-0002-1983-481X
Researcher of the Laboratory of Respiratory Viral Infection with Drug Testing
Russian Federation, 123098, MoscowLidya B. Kisteneva
N.F. Gamaleya National Research Center of Epidemiology and Microbiology of the Ministry of Health of Russia
Email: lborisovna2007@yandex.ru
ORCID iD: 0000-0001-7336-409X
D. Sci. in Med., Leading Researcher of the Laboratory of Chronic Viral Infections
Russian Federation, 123098, MoscowLyudmila V. Kolobukhina
N.F. Gamaleya National Research Center of Epidemiology and Microbiology of the Ministry of Health of Russia
Email: lkolobuchina@yandex.ru
ORCID iD: 0000-0001-5775-3343
D. Sci. in Med., Chief Researcher of Respiratory Viral Infection with Drug Testing
Russian Federation, 123098, MoscowElena I. Burtseva
N.F. Gamaleya National Research Center of Epidemiology and Microbiology of the Ministry of Health of Russia
Email: elena-burtseva@yandex.ru
ORCID iD: 0000-0003-2518-6801
D. Sci. in Med., Leading Researcher of the Influenza Etiology and Epidemiology
Russian Federation, 123098, MoscowReferences
- Iuliano A.D., Roguski K.M., Chang H.H., Muscatello D.J., Palekar R., Tempia S., et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2018; 391(10127): 1285–300. https://doi.org/10.1016/s0140-6736(17)33293-2
- Аcute respiratory viral diseases in adults: Clinical recommendations. National Infectious Diseases Scientific Society, 2014. Available at: https://library.mededtech.ru/rest/documents/ORVI_adult/ (in Russian)
- Isakov V.A. Clinical and pathogenetic aspects of severe influenza. Allergologiya i immunologiya. 2002; 3(1): 136–44. https://elibrary.ru/xvrzmd (in Russian)
- Pokrovskii V.I., Semenov B.F. The concept of delayed death in influenza and vaccine prevention tactics for heart attacks, strokes and deaths from this infection. Rossiiskii meditsinskii zhurnal. 2003; 11(22): 1266–72. https://elibrary.ru/weoptp (in Russian)
- Shi J., Zeng X., Cui P., Yan C., Chen H. Alarming situation of emerging H5 and H7 avian influenza and effective control strategies. Emerg. Microbes Infect. 2023; 12(1): 2155072. https://doi.org/10.1080/22221751.2022.2155072
- de Jong M.D., Simmons C.P., Thanh T.T., Hien V.M., Smith G.J., Chau T.N., et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 2006; 12(10): 1203–7. https://doi.org/10.1038/nm1477
- Nikolaeva L.I., Sapronov G.V., Kupriyanov V.V. The role of the interferons-lambda in immune protection against influenza. Infektsionnye bolezni. 2019; 17(1): 86–92. https://doi.org/10.20953/1729-9225-2019-1-86-92 https://elibrary.ru/dcfhfp (in Russian)
- Klinkhammer J., Schnepf D., Ye L., Schwaderlapp M., Gad H.H., Hartmann R., et al. IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. Elife. 2018; 7: e33354. https://doi.org/10.7554/elife.33354
- Fox J.M., Crabtree J.M., Sage L.K., Tompkins S.M., Tripp R.A. Interferon lambda upregulates IDO1 expression in respiratory epithelial cells after influenza virus infection. J. Interferon Cytokine Res. 2015; 35(7): 554–62. https://doi.org/10.1089/jir.2014.0052
- Davidson S., McCabe T.M., Crotta S., Gad H.H., Hessel E.M., Beinke S., et al. IFNλ is a potent anti-influenza therapeutic without the inflammatory side effects of IFNα treatment. EMBO Mol. Med. 2016; 8(9): 1099–112. https://doi.org/10.15252/emmm.201606413
- Kim S., Kim M.J., Kim C.H., Kang J.W., Shin H.K., Kim D.Y., et al. The superiority of IFN-λ as a therapeutic candidate to control acute influenza viral lung infection. Am. J. Respir. Cell Mol. Biol. 2017; 56(2): 202–12. https://doi.org/10.1165/rcmb.2016-0174oc
- Feld J.J., Kandel C., Biondi M.J., Kozak R.A., Zahoor M.A., Lemieux C., et al. Peginterferon lambda for the treatment of outpatients with COVID-19: a phase 2, placebo-controlled randomised trial. Lancet Respir. Med. 2021; 9(5): 498–510. https://doi.org/10.1016/s2213-2600(20)30566-x
- Lazear H.M., Nice T.J., Diamond M.S. Interferon-λ: immune functions at barrier surfaces and beyond. Immunity. 2015; 43(1): 15–28. https://doi.org/10.1016/j.immuni.2015.07.001
- Otdel’nova K.A. Determination of the required number of observations in complex social and hygienic studies. In: Proceedings of N.I. Pirogov Medical Institute «Complex socio-hygienic studies and clinical and social research» [Sbornik trudov meditsinskogo instituta im. N.I. Pirogova «Kompleksnye sotsial’no-gigienicheskie issledovaniya i kliniko-sotsial’nye issledovaniya»]. Moscow; 1980. (in Russian)
- L’vov D.K., Burtseva E.I., Kolobukhina L.V., Fedyakina I.T., Kirillova E.S., Trushakova S.V., et al. Virological, epidemiological, clinic, and molecular genetic features of the influenza epidemic in 2015-2016: prevailing of the influenza A(H1N1)09 pdm virus in Russia and countries of the Northern hemisphere. Voprosy virusologii. 2016; 61(4): 159–65. https://doi.org/10.18821/0507-4088-2016-61-4-159-166 (in Russian)
- Kurdin A., Ambalov Yu., Pshenichnaya N. The importance of polymorphisms of lambda-interferon genes in pathogenesis and clinical aspects of Flua and other acute respiratory-viral infections. Glavnyi vrach Yuga Rossii. 2017; (3): 49–50. https://elibrary.ru/yzbirt (in Russian)
- Keshavarz M., Namdari H., Farahmand M., Mehrbod P., Mokhtari-Azad T., Rezaei F. Association of polymorphisms in inflammatory cytokines encoding genes with severe cases of influenza A/H1N1 and B in an Iranian population. Virol. J. 2019; 16(1): 79. https://doi.org/10.1186/s12985-019-1187-8
- Parakhonskii A.P. Aging of the immune system. Mezhdunarodnyi zhurnal prikladnykh i fundamental’nykh issledovanii. 2011; (6-1): 73–4. https://elibrary.ru/necmtj (in Russian)
Supplementary files