Genetic diversity of the human immunodeficiency virus (HIV-1) in the Kaliningrad region
- Authors: Shchemelev A.N.1, Semenov A.V.2, Ostankova Y.V.1, Naidenova E.V.3, Zueva E.B.1, Valutite D.E.1, Churina M.A.4, Virolainen P.A.1, Totolian A.A.1
-
Affiliations:
- FBSI «Saint Petersburg Pasteur Research Institute of Epidemiology and Microbiology» of the Federal Service for Surveillance of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
- Ekaterinburg Research Institute of Viral Infections of the Federal Research Institute, State Research Center for Virology and Biotechnology “Vector” of the Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
- FSSI Russian Research Anti-Plague Institute «Microbe» of the Federal Service for Surveillance of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
- St. Petersburg GBUZ «Botkin Clinical Infectious Diseases Hospital»
- Issue: Vol 67, No 4 (2022)
- Pages: 310-321
- Section: ORIGINAL RESEARCH
- URL: https://virusjour.crie.ru/jour/article/view/633
- DOI: https://doi.org/10.36233/0507-4088-119
- ID: 633
Cite item
Abstract
Introduction. As is currently known, the epidemic process in the Kaliningrad Region was mainly associated with the spread of the recombinant form of HIV-1 (CRF03_AB); however, regular HIV importations from other countries and continents has created favorable conditions for emergence and spread of various recombinant forms of the virus.
The most complete information on the diversity of recombinant forms in the region is also necessary to understand the structure of drug resistance (DR).
The aim of the study was to explore the HIV-1 genetic diversity in the Kaliningrad Region.
Materials and methods. We studied 162 blood plasma samples obtained from patients from the Kaliningrad Region, both with confirmed virological failure of antiretroviral therapy (ART) and with newly diagnosed HIV infection. For reverse transcription and amplification of HIV genome fragments, diagnostic «AmpliSense HIVResist-Seq».
Results and discussion. The various recombinants between subtypes A and B (74%) were predominant in study group: recombinant was between CRF03_AB and subtype A (33.95%) and CRF03_AB-like (13.58%) were the most common. Among the “pure” subtypes of the virus, subtype A6 (16.67%). The circulation of subtypes B (3.70%) and G (1.23%) was also noted.
Ninety-six patients (59.26%) were identified with at least one mutation associated with antiretroviral (ARV) drug resistance.
Conclusion. The observed diversity of subtypes and recombinant forms of the virus implies that the new recombinants are actively emerging in the studied region, both between existing recombinant forms and “pure” subtypes, as well as between “pure” subtypes.
Full Text
Introduction
The human immunodeficiency virus type 1 (HIV-1) was first isolated in the 1980s and has been circulating in the human population for almost 100 years. The stable phylogenetic structure that has been developed over that time is represented by three main groups: N, O, M; the latter, in its turn, is divided into 12 distinct subtypes comprising viral isolates that are more closely related to each other than to isolates from other subtypes [1, 2]. Classification of HIV-1 was originally based on sequences of subgenomic regions or individual genes; however, recent improvements in sequencing methods made it possible to classify HIV-1 based on full-length genomes or sequences from multiple subgenomic regions. As a result, isolates could be identified with distinctive parts of their genomes corresponding to different subtypes: These isolates are products of recombination between parental strains belonging to different subtypes. When a particular recombinant form is identified in three or more individuals with no direct epidemiological linkage, it is classified as a circulating recombinant form (CRF). At present, there are more than one hundred known CRFs [3] being responsible for at least 20% of global HIV epidemics [4], with recombinant forms prevailing in several regions such as Western and Central Africa (CRF02_AG) [5, 6] and Southeast Asia (CRF01_AE) [7, 8].
Recombinant forms of the virus are the product of recombination during reverse transcription. During the minus-strand deoxyribonucleic acid (DNA) synthesis, the reverse transcriptase (RT) shifts from one ribonucleic acid (RNA) strand to the other with a high frequency, suggesting that at least both genomic RNA copies are used alternatively as templates. The copy choice switching rate in HIV-1 for highly similar template RNAs is estimated at 3 × 10–4 – 1.4 × 10–3 events per nucleotide, i.e. 3–12 template switches per genome replication [9–11]. Importantly, the formation of virions containing two different genomic RNAs depends on fulfilment of certain prerequisites: First, two or more viruses with different genotypes must infect the same cell; second, genomic RNAs of different origin must be subsequently co-packaged. Such situation can be associated either with coinfection or superinfection of a patient with different virus subtypes.
The fact that the Nef and Vpu proteins downregulate the expression of CD4 and co-receptors during HIV-1 infection implies that superinfections are of extremely rare occurrence [12]. Nevertheless, the in-situ hybridization of cells from patients showed that single cells could contain more than four different proviruses [13, 14]. In addition, the high recombination rates and spread of recombination forms demonstrated by recent studies imply high rates of coinfection in vivo [4, 15]. Thus, different HIV subtypes must co-circulate in a region to activate the generation of recombinant forms of the virus.
In the Russian Federation, the most prevalent sub-subtype of the virus is A6 also known as IDU-A (Injecting Drug Users) or A-FSU (former Soviet Union countries). This sub-subtype was previously classified as A1; however, being significantly different from other HIV-1 variants of sub-subtype A1 by its structure and transmission, it was set apart as an individual, relatively uniform group [16, 17]. In the meantime, some regions have become favorable for co-circulation of several subtypes, including the Kaliningrad Region and its central city, which is a major transportation hub with railroads and highways, sea and river ports, and the international airport.
As is currently known, the epidemic process the Kaliningrad Region was initially associated with the spread of the recombinant form of the virus (CRF03_AB) among injecting drug users. Later, the HIV infection expanded beyond the vulnerable groups of population. In addition, regular importation of HIV from other countries and continents has created favorable conditions for emergence of various new recombinant forms of the virus in the region [18, 19].
Notably, the spread of subtypes and recombinant forms in the HIV-1 epidemic is very dynamic: the contemporary virus genetic diversity is represented by a mixture of recombinants, which emerged during earlier stages of the global epidemic, and recombinants, which emerged later; all of them are contributing to creation of more complex recombinant forms, which subsequently will make their contribution to the dynamics of the HIV-1 global population. It can be assumed that the process of generation of increasingly complex recombinant forms will constitute the core of the virus evolution in the Kaliningrad Region.
The aim of the study was to explore the HIV-1 genetic diversity in the Kaliningrad Region.
Materials and methods
The study was performed in 2014–2018 using clinical material from 162 patients from the Kaliningrad Region, both with confirmed virological failure on antiretroviral therapy (ART) and with newly diagnosed HIV infection. To detect HIV resistant strains, the blood plasma was delivered to the North-West District Center for AIDS Prevention and Control (NWD AIDS Center) of the St. Petersburg Pasteur Research Institute of Epidemiology and Microbiology.
The blood plasma was used for measuring the viral load with the AmpliSens HIV-Monitor-FRT reagent kit (the Central Research Institute of Epidemiology, Russia) with the sensitivity threshold of 500 copies/ml. The samples with detectable viral load were further tested through reverse transcription polymerase chain reaction (RT-PCR) and Sanger sequencing. Diagnostic RT-PCR-kit-Pro/Rev and PCR-kit-Pro/Rev kits (the Central Research Institute of Epidemiology, Russia) were used for HIV reverse transcription and amplification; the sequencing reaction was performed in accordance with the manual of the AmpliSens HIVResist-Seq kit (Central Research Institute of Epidemiology, Russia). The genotyping of HIV-1 was based on the analysis of nucleotide sequences of the pol gene fragment of 1,302 nt, encoding protease (PR) and part of reverse transcriptase (RT) in the 2,253–3,554 nt region; the coordinates are given for HIV HXB2 (GenBank accession number K03455.1) in the international GenBank database. The products of the sequencing reaction were analyzed using the ABI Prism 3500 genetic analyzer (Applied Biosystems, United States).
The primary analysis of nucleotide sequences was performed using the NCBI Blast program to compare with nucleotide sequences available in the international GenBank database. The alignment of nucleotide sequences was performed using the MEGA 7.0 software and the ClustalW algorithm [20]. For phylogenetic trees and for the subsequent phylogenetic analysis, we used the Neighbor-Joining algorithm to optimize trees in accordance with the balanced minimum evolution criterion. To assess the significance of phylogenetic relationships, we used the bootstrap method to generate multiple samples for 1,000 independent constructions of each phylogenetic tree.
The genotyping of studied isolates was performed using the REGA HIV-1 Subtyping Tool 3.0 software [21] and the analysis of their phylogenetic relationships with reference sequences from the international GenBank database. To identify and analyze recombinant forms, we used the REGA HIV-1 Subtyping Tool 3.0 software and the parameters preset in the software (window size – 400 bp; step size – 20). The HIV-1 genetic sequences were assessed for drug-resistance (DR) mutations using the Stanford database (Stanford HIV DB) [22]. The mutational profiles were analyzed by building linear diagrams using the Linear Diagram Generator software [23].
The statistical analysis of the data was performed using MS Excel Professional Plus 2013 (Microsoft), Prizm v5.0 (GraphPad Software Inc.) software. The statistical error was measured using the Clopper-Pierson exact interval. The results were presented with a 95% confidence interval (CI). To measure the significance of differences between numerical data obtained in paired comparisons, we used (depending on the characteristics of samples) Fisher’s exact test or the χ2 test with Yates’ correction. The probability value as the significance threshold for differences was set at p < 0.05.
The study was performed with the informed consent of the patients. The research protocol was approved by the Ethics Committee of the St. Petersburg Pasteur Research Institute of Epidemiology and Microbiology (Minutes No. 3 of 7/4/2010 and Minutes No. 47 of 25/12/2018).
Results and discussion
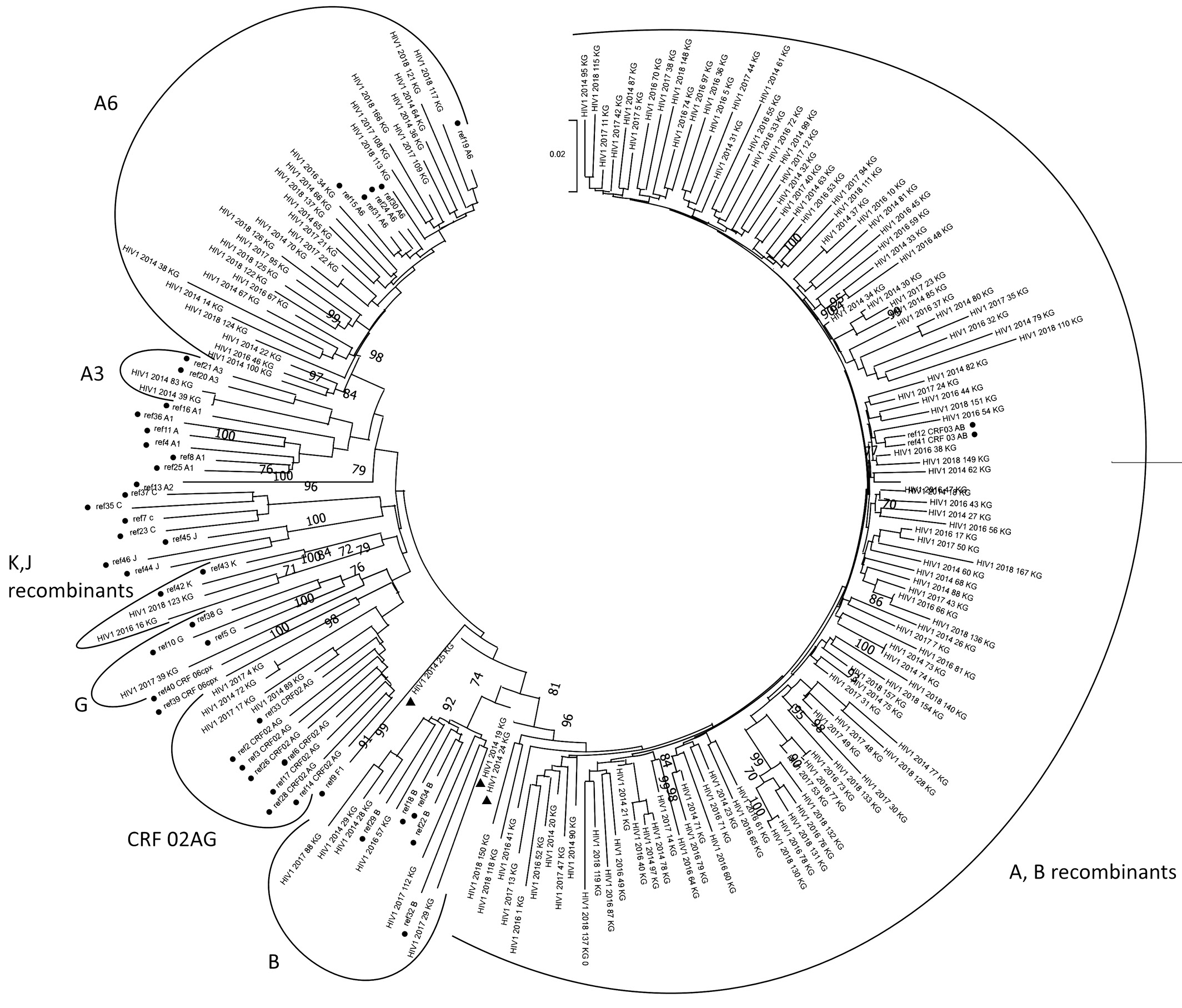
The sequences of 162 HIV-1 isolates were obtained to be further deposited to GenBank under numbers ON367567–ON367728. The sub-subtype was identified for all of them (Table 1). Both the data obtained during the genotyping using the REGA HIV Subtyping Tool 3.0 and jumping profile Hidden Markov Model (jpHMM) (Supplement А) and the results of the phylogenetic analysis conducted using the Mega X software (Fig. 1) were taken into consideration.
Table 1. Distribution of isolates by HIV-1 subtypes
Таблица 1. Распределение исследованных штаммов по субтипам ВИЧ-1
Subtype Субтип | Number of isolates Количество штаммов | Sample Share, % Доля в выборке, % | 95% CI, % 95% ДИ, % | |
– | + | |||
HIV-1 Subtype A3 | 2 | 1,23 | 0,15 | 4,39 |
HIV-1 Subtype A6 | 27 | 16,67 | 11,28 | 23,31 |
HIV-1 Subtype B | 6 | 3,70 | 1,37 | 7,89 |
HIV-1 Subtype G | 2 | 1,23 | 0,15 | 4,39 |
HIV-1 CRF03_AB | 41 | 25,31 | 18,81 | 32,73 |
HIV-1 CRF03_AB-like | 22 | 13,58 | 8,71 | 19,84 |
Recombinant of 03_AB, A | 55 | 33,95 | 26,71 | 41,79 |
HIV-1 CRF02_AG | 4 | 2,47 | 0,68 | 6,20 |
Recombinant of A1, B | 2 | 1,23 | 0,15 | 4,39 |
Recombinant of K, J | 2 | 1,23 | 0,15 | 4,39 |
Fig. 1. Results of phylogenetic analysis using the Neighbor Joining algorithm. • – reference sequences (table 2); ▲ – recombinant forms between subtypes A and B, not clustered with other recombinants of this group.
The dominance of recombinants between subtypes A and B in the region, including CRF03_AB, is supported by the published data on the genetic diversity of the virus in the Kaliningrad Region [16, 18, 19]. Nevertheless, the heterogeneity of recombinants inside the clade in the phylogenetic tree is of interest, being associated with characteristics of epidemiological relationships and new circulating recombinant forms emerging in the region. It requires further studies on full-length genomes of virus isolates from the Kaliningrad Region.
Among recombinants between subtypes A and B, special attention should be given to three isolates marked in the phylogenetic tree (Fig. 1). In the dendrogram, recombinants between subtypes A and B form a large heterogenous cluster, while the above isolates are separated from them, although their pol gene has fragments related to subtypes A and B, as confirmed by the comparative recombination analysis using different software (Fig. 2). At the same time, their subtyping with different online tools (REGA, Geno2Pheno, NCBI, Stanford HIV DB, RIP) precludes from making the conclusion about the genotypic affinity of these isolates. Note that all the three specimens have different location in the dendrogram: isolate HIV1_2014_24_KG is closest to other recombinant forms between subtypes A and B; isolate HIV1_2014_19_KG clusters with subtype B isolates; isolate HIV1_2014_25_KG forms the earliest branch – at the level of divergence of recombinants A and B as well as other HIV-1 subtypes. All the three isolates were collected from patients who were infected relatively recently (less than one year ago) and were not administered ART. The analysis of genetic sequences in chromatograms revealed multiple degenerated fragments, i.e. presence of several different nucleotides at the same positions in the genome. As known, the observed situation can be indicative of the diversity of the viral population in the patient, including coinfection with different HIV subtypes [24]. Thus, it can be assumed that the above isolates are at the beginning of the retroviral recombination process, being primarily represented by virus variants with genomes of A and B isolates co-packaged in the capsid rather than by recombinant forms.
Fig. 2. Comparative recombination analysis of samples 2014_80 (CRF03_AB) and 2014_19 (A + B recombinant) in Rega HIV Subtyping Tool v3.0 [21] and Recombinant Identification Program (https://www.hiv.lanl.gov/content/sequence/RIP/RIP.html) a – sample 2014_80 in Rega HIV Subtyping Tool v3.0; b – sample 2014_80 in the Recombinant Identification Program; c – sample 2014_19 in Rega HIV Subtyping Tool v3.0; d – sample 2014_19 in the Recombinant Identification Program [25].
Table 2. Names of reference sequences from GenBank used in phylogenetic analysis
Таблица 2. Наименование референсных последовательностей из GenBank, использованных в филогенетическом анализе
Sub- subtype Суб- субтип | Number sequence in phylogenetic tree Номер последовательности на филогенетическом древе | Number sequence in GenBank Номер последовательности из GenBank | Region of origin Регион происхождения |
A1 | ref4 | AF069670 | Somali Сомали |
A1 | ref8 | AB287376 | Ruanda Руанда |
A1 | ref16 | U51190 | Uganda Уганда |
A1 | ref25 | EU110087 | Kenia Кения |
A1 | ref27 | AF484509 | Uganda Уганда |
A1 | ref36 | AF107771 | Sweden Швеция |
A2 | ref13 | AF286237 | Cyprus Кипр |
A3 | ref1 | AY521631 | Senegal Сенегал |
A3 | ref20 | AY521629 | Sweden Швеция |
A6 | ref15 | HQ449397 | Russia, Krasnodar Россия, Краснодар |
A6 | ref19 | HQ161930 | Russia, Smolensk Россия, Смоленск |
A6 | ref24 | EF589043 | Kazakhstan Казахстан |
A6 | ref30 | AY500393 | Russia, Moscow Россия, Москва |
A6 | ref31 | AF413987 | Ukraine Украина |
B | ref18 | M17449 | USA США |
B | ref22 | KJ771697 | Germany Германия |
B | ref29 | HM586190 | Great Britain Великобритания |
B | ref32 | AY713409 | USA США |
B | ref34 | AY173951 | Thailand Таиланд |
C | ref7 | AF067155 | India Индия |
C | ref23 | U52953 | Brazil Бразилия |
C | ref35 | U46016 | Ethiopia Эфиопия |
C | ref37 | AY772699 | Africa Африка |
F1 | ref9 | AF075703 | Finland Финляндия |
G | ref5 | AF061641 | Finland Финляндия |
G | ref10 | U88826 | Nigeria Нигерия |
G | ref38 | AF084936 | Congo Конго |
J | ref44 | EF614151 | Congo Конго |
J | ref45 | GU237072 | Cameron Камерун |
J | ref46 | AF082394 | Sweden Швеция |
К | ref42 | AJ249235 | Cameron Камерун |
К | ref43 | AJ249239 | Cameron Камерун |
CRF02_AG | ref2 | AF063224 | Djibouti Джибути |
CRF02_AG | ref3 | GU201514 | Cameron Камерун |
CRF02_AG | ref6 | KT124792 | Germany Германия |
CRF02_AG | ref14 | AB231898 | Ghana Гана |
CRF02_AG | ref17 | EU786671 | Spain Испания |
CRF02_AG | ref26 | AB231896 | Ghana Гана |
CRF02_AG | ref28 | AY151001 | Ecuador Эквадор |
CRF02_AG | ref33 | AF377954 | Cameron Камерун |
CRF06_cpx | ref39 | HQ529257.1 | Ghana Гана |
CRF03_AB | ref12 | AF193276 | Russia, Kalinigrad Россия, Калининград |
CRF03_AB | ref41 | AF414006.1 | Belarus Беларусь |
CRF06_cpx | ref40 | MH605500.1 | Guinea-Bissau Гвинея-Бисау |
In addition to recombinants between subtypes A and B, the study has revealed CRF02_AG CRFs, which are of rare occurrence in the European part of Russia, and recombinants between subtypes K and J [26].
Among the “pure” subtypes of the virus, the leading place is taken by subtype A common in Russia and represented by two sub-subtypes – A6 (16.67%; 95% CI 11.28–23.31%) and A3 (1.23%; 95% CI 0.15–4.39%); the co-circulating subtypes are subtypes B (3.70%; 95% CI 1.37–7.89%) and G (1.23%; 95% CI 0.15–4.39%).
The studied region demonstrates the distribution of HIV-1 subtypes, which is different from other regions of Russia in general and the North-West Federal District in particular [27–29]. To compare the significance of differences in the genetic diversity among regions of the North-West Federal District, we selected sub-subtype A6, subtype B, and recombinant forms between subtypes A and B, as they have been detected not only in the samples analyzed in this study, but also in isolates from Arkhangelsk [28] and Leningrad Regions [29]. The significance of differences was assessed using the χ2 test with Yates’ correction. No significant differences between the occurrence frequencies of HIV-1 subtypes in Arkhangelsk and Leningrad Regions were found, though statistically significant differences in the genetic diversity were demonstrated by the above regions and the Kaliningrad Region (χ2 is 254.277; the critical value of χ2 is 13.277 at the significance level p = 0.01).
The differences in the genetic diversity result from the dominance of HIV-1 recombinant forms in the Kaliningrad Region, while they were of rare occurrence in Arkhangelsk and Leningrad Regions. At the same time, the diversity of “pure” subtypes of HIV-1 matches the diversity described in published studies [27–29], with dominating subtype A, primarily sub-subtype A6.
The observed dominance of virus variants, which are recombinants between CRF03_AB and subtype A, as well as the dominance of the recombinant form similar to CRF03_AB, though having a number of differences (CRF03_AB-like), support the assumption stating that the long-lasting co-circulation of recombinant forms and “pure” subtypes of the virus results in emergence of new, more complex recombinant forms and new fragments included in the genome [4].
In addition to the genotypic analysis, we analyzed the frequency of DR associated mutations in this region. The analysis included isolates collected from patients with ART failure (n = 107) and from patients with newly diagnosed infection (n = 55). The primary DR was detected only in two cases (3.64%; 95% CI 0.44–12.53%); therefore, the analysis includes all the patients with detected DR mutations.
A total of 80 different DR associated mutations were detected. Most of them were DR mutations to RT inhibitors, including nucleoside reverse transcriptase inhibitors (NRTIs) – 31 substitutions (38.75%; 95% CI 28.06–50.30%) and non-nucleoside reverse transcriptase inhibitors (NNRTIs) – 35 mutations (43.75%; 95% CI 32.68–55.30%); substitutions associated with DR to protease inhibitors (PIs) – 14 (17.50%; 95% CI 9.91–27.62%) account for a lesser proportion of mutational diversity.
96 patients (59.26%; 95% CI 51.27–66.90%) had HIV-1 isolates with at least one mutation associated with DR to antiretroviral agents. The most frequently detected mutations were DR mutations to RT inhibitors. In 13 cases, we detected DR mutations to NRTIs; 4 patients had DR mutations to NNRTIs and 66 had DR mutations to NRTIs + NNRTIs. In addition, 13 patients had DR mutations to PIs: 10 patients – to PIs + NRTIs, and 3 patients – to PIs + NRTIs + NNRTIs.
Among DR mutations to NRTIs, the highest frequency rates were demonstrated by M184V mutations [30] (65.63%; 95% CI 55.23–75.02%), L74V mutations [31] (19.79%; 95% CI 12.36–29.17%), Y115F mutations [32] (14.58%; 95% CI 8.21–23.26%); the other substitutions were detected in 10% of the cases and more rarely. The analysis of multiple mutational profiles by building linear diagrams showed stable patterns of DR mutations (Fig. 3 a). The thymidine analog resistance mutations (TAMs) extensively described in the scientific publications were detected in the obtained profiles only in a few cases. There are two pathways of development of ТАМ patterns: mutations occurring with T215Y (including M41L, L210W and sometimes D67N) comprise the TAM-1 cluster; mutations occurring with K70R (including D67N, T215F, and K219Q) comprise the TAM-2 cluster. Nevertheless, in this case, both mutation clusters are associated with the T215Y substitution, while the development of the pattern along the TAM-2 pathway is known to have the greatest advantage with the T215F substitution, which was also detected in the studied mutational profiles, but not in the TAM patterns [33]. The profiles carrying non-TAM mutations prevailed; they have demonstrated a stable relationship between L74V + Y115F substitutions. These mutations are primarily associated with DR to abacavir and didanosine, though there are data on their association with DR to tenofovir [31, 32], which, in its turn, is included in most of the present-day antiretroviral treatment regimens. Furthermore, in all cases, this combination was detected together with the M184V substitution, which most likely can be explained by the presence of this substitution in most isolates with DR.
Fig. 3. Results of the study of multiple mutational profiles by constructing line diagrams: a – for NRTI resistance mutations; b – for NNRTI resistance mutations.
The analysis of frequency rates for DR mutations to NNRTIs showed that the highest frequency rates were demonstrated by the K103N [34] (36.46%; 95% CI 26.87–46.91%), K101E [35] (12.50%; 95% CI 6.63–20.82%), G190A [36] (11.46%; 95% CI 5.86–19.58%), P225H (15.63%; 95% CI 9.02–24.46%), and Y18C [37] (12.50%; 95% CI 6.63–20.82%) substitutions; the other mutations were detected in less than 10% of the cases. The analysis of profiles of DR mutations in obtained isolates (Fig. 3 b) revealed a relationship between K101E + G190A/S substitutions; note that this combination was generally detected without the most common K103N mutation. The analysis also revealed a relationship between the substitution for alanine (A) or serine (S) at the 190th position and the subtype of the virus. Substitution 190A was detected only in recombinants between subtypes A and B, while mutation 190S was detected primarily in isolates of sub-subtype A6 (in five of six cases). There are published studies describing the prevalence of the substitution at the 190th position of RT for serine for subtype A [38–40] and alanine for non-A subtypes [38, 41, 42].
Conclusion
The obtained results demonstrate the existence of diverse recombinant forms in the Kaliningrad Region. The dominance of recombinants between CRF03_AB and A suggests that the recombination generally results from the co-circulation of the variant common in the studied region – CRF03_AB, and sub-subtype A6 common in other regions of Russia. The contribution of the co-circulation with subtype B remains unclear.
The observed diversity of subtypes and recombinant forms of the virus implies that new recombinants are actively emerging in the studied region, both between the existing recombinant forms and “pure” subtypes as well as between “pure” subtypes. This activity of the virus adds to the importance of studies on full-length genomes of the isolates obtained in the Kaliningrad Region and of the description of all the recombinant forms currently circulating in the region.
About the authors
Alexander N. Shchemelev
FBSI «Saint Petersburg Pasteur Research Institute of Epidemiology and Microbiology» of the Federal Service for Surveillance of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
Author for correspondence.
Email: tvildorm@gmail.com
ORCID iD: 0000-0002-3139-3674
Junior Researcher, Laboratory of Immunology and Virology of HIV Infection, St. Petersburg Pasteur Research Institute of Epidemiology and Microbiology, Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
Russian Federation, 197101, St. PetersburgAleksandr V. Semenov
Ekaterinburg Research Institute of Viral Infections of the Federal Research Institute, State Research Center for Virology and Biotechnology “Vector” of the Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
Email: tvildorm@gmail.com
ORCID iD: 0000-0003-3223-8219
Russian Federation, 620030 Ekaterinburg
Yulia V. Ostankova
FBSI «Saint Petersburg Pasteur Research Institute of Epidemiology and Microbiology» of the Federal Service for Surveillance of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
Email: tvildorm@gmail.com
ORCID iD: 0000-0003-2270-8897
Russian Federation, 197101, St. Petersburg
Ekaterina V. Naidenova
FSSI Russian Research Anti-Plague Institute «Microbe» of the Federal Service for Surveillance of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
Email: tvildorm@gmail.com
ORCID iD: 0000-0001-6474-3696
Russian Federation, 410005, Saratov
Elena B. Zueva
FBSI «Saint Petersburg Pasteur Research Institute of Epidemiology and Microbiology» of the Federal Service for Surveillance of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
Email: tvildorm@gmail.com
ORCID iD: 0000-0002-0579-110X
Russian Federation, 197101, St. Petersburg
Diana E. Valutite
FBSI «Saint Petersburg Pasteur Research Institute of Epidemiology and Microbiology» of the Federal Service for Surveillance of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
Email: tvildorm@gmail.com
ORCID iD: 0000-0002-0931-102X
Russian Federation, 197101, St. Petersburg
Mariia A. Churina
St. Petersburg GBUZ «Botkin Clinical Infectious Diseases Hospital»
Email: tvildorm@gmail.com
ORCID iD: 0000-0003-0424-4654
Russian Federation, 191167, St. Petersburg
Pavel A. Virolainen
FBSI «Saint Petersburg Pasteur Research Institute of Epidemiology and Microbiology» of the Federal Service for Surveillance of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
Email: tvildorm@gmail.com
ORCID iD: 0000-0001-5918-9395
Russian Federation, 197101, St. Petersburg
Areg A. Totolian
FBSI «Saint Petersburg Pasteur Research Institute of Epidemiology and Microbiology» of the Federal Service for Surveillance of Consumer Rights Protection and Human Welfare (Rospotrebnadzor)
Email: tvildorm@gmail.com
ORCID iD: 0000-0003-4571-8799
Russian Federation, 197101, St. Petersburg
References
- Korber B., Muldoon M., Theiler J., Gao F., Gupta R., Lapedes A., et al. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000; 288(5472): 1789–96. https://doi.org/10.1126/science.288.5472.1789
- Kuiken C., Foley B., Hahn B., Marx P., McCutchan F., Mellors J.W., et al. A compilation and analysis of nucleic acid and amino acid sequences. In: Human Retroviruses and AIDS. Los Alamos; 1999.
- Los Alamos National Laboratory. HIV Circulating Recombinant Forms (CRFs). Available at: https://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html
- Simon-Loriere E., Rossolillo P., Negroni M. RNA structures, genomic organization and selection of recombinant HIV. RNA Biol. 2011; 8(2): 280–6. https://doi.org/10.4161/rna.8.2.15193
- McCutchan F.E., Carr J.K., Bajani M., Sanders-Buell E., Harry T.O., Stoeckli T.C., et al. Subtype G and multiple forms of A/G intersubtype recombinant human immunodeficiency virus type 1 in Nigeria. Virology. 1999; 254(2): 226–34. https://doi.org/10.1006/viro.1998.9505
- Montavon C., Toure-Kane C., Liegeois F., Mpoudi E., Bourgeois A., Vergne L., et al. Most env and gag subtype A HIV-1 viruses circulating in West and West Central Africa are similar to the prototype AG recombinant virus IBNG. J. Acquir. Immune. Defic. Syndr. 2000; 23(5): 363–74. https://doi.org/10.1097/00126334-200004150-00001
- Menu E., Truong T.X., Lafon M.E., Nguyen T.H., Müller-Trutwin M.C., Nguyen T.T., et al. HIV type 1 Thai subtype E is predominant in South Vietnam. AIDS Res. Hum. Retroviruses. 1996; 12(7): 629–33. https://doi.org/10.1089/aid.1996.12.629
- Piyasirisilp S., McCutchan F.E., Carr J.K., Sanders-Buell E., Liu W., Chen J., et al. A recent outbreak of human immunodeficiency virus type 1 infection in southern China was initiated by two highly homogeneous, geographically separated strains, circulating recombinant form AE and a novel BC recombinant. J. Virol. 2000; 74(23): 11286–95. https://doi.org/10.1128/jvi.74.23.11286-11295.2000
- Galetto R., Moumen A., Giacomoni V., Veron M., Charneau P., Negroni M. The structure of HIV-1 genomic RNA in the gp120 gene determines a recombination hot spot in vivo. J. Biol. Chem. 2004; 279(35): 36625–32. https://doi.org/10.1074/jbc.m405476200
- Zhuang J., Jetzt A.E., Sun G., Yu H., Klarmann G., Ron Y., et al. Human immunodeficiency virus type 1 recom-bination: rate, fidelity and putative hot spots. J. Virol. 2002; 76(22): 11273–82. https://doi.org/10.1128/jvi.76.22.11273-11282.2002
- Jetzt A.E., Yu H., Klarmann G.J., Ron Y., Preston B.D., Dougherty J.P. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 2000; 74(3): 1234–40. https://doi.org/10.1128/jvi.74.3.1234-1240.2000
- Piantadosi A., Chohan B., Chohan V., McClelland R.S., Overbaugh J. Chronic HIV-1 infection frequently fails to protect against superinfection. PLoS Pathog 2007; 3(11): 177. https://doi.org/10.1371/journal.ppat.0030177
- Gratton S., Cheynier R., Dumaurier M.J., Oksenhendler E., Wain-Hobson S. Highly restricted spread of HIV-1 and multiply infected cells within splenic germinal centers. Proc. Natl. Acad. Sci. USA. 2000; 97(26): 14566–71. https://doi.org/10.1073/pnas.97.26.14566
- Jung A., Maier R., Vartanian J.P., Bocharov G., Jung V., Fischer U., et al. Recombination: Multiply infected spleen cells in HIV patients. Nature. 2002; 418(6894): 144. https://doi.org/10.1038/418144a
- Chen J., Dang Q., Unutmaz D., Pathak V.K., Maldarelli F., Powell D., et al. Mechanisms of nonrandom human immunodeficiency virus type 1 infection and double infection: preference in virus entry is important but is not the sole factor. J. Virol 2005; 79(7): 4140–9. https://doi.org/10.1128/jvi.79.7.4140-4149.2005
- Bobkov A.F., Kazennova E.V., Selimova L.M., Khanina T.A., Ryabov G.S., Bobkova M.R., et al. Temporal trends in the HIV-1 epidemic in Russia: predominance of subtype A. J. Med. Virol. 2004; 74(2): 191–6. https://doi.org/10.1002/jmv.20177
- Schlösser M., Kartashev V.V., Mikkola V.H., Shemshura A., Saukhat S., Kolpakov D., et al. HIV-1 sub-subtype A6: Settings for normalised identification and molecular epidemiology in the Southern Federal District, Russia. Viruses. 2020; 12(4): 475. https://doi.org/10.3390/v12040475
- Liitsola K., Tashkinova I., Laukkanen T., Korovina G., Smolskaja T., Momot O., et al. HIV-1 genetic subtype A/B recombinant strain causing an explosive epidemic in injecting drug users in Kaliningrad. AIDS. 1998; 12(14): 1907–19. https://doi.org/10.1097/00002030-199814000-00023
- Lebedev A., Pasechnik O., Ozhmegova E., Antonova A., Blokh A., Grezina L., et al. Prevalence and spatiotemporal dynamics of HIV-1 Circulating Recombinant Form 03_AB (CRF03_AB) in the Former Soviet Union countries. PLoS One. 2020; 15(10): e0241269. https://doi.org/10.1371/journal.pone.0241269
- Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016; 33(7): 1870–4. https://doi.org/10.1093/molbev/msw054
- Stanford University. HIV Drug Resistance Database. REGA HIV-1 Subtyping Tool – Version 3.0. Available at: http://dbpartners.stanford.edu:8080/RegaSubtyping/stanford-hiv/typingtool/
- Stanford University. HIV Drug Resistance Database. HIVdb Program: Mutations Analysis. Available at: https://hivdb.stanford.edu/hivdb/by-patterns/
- Gottfried B. A comparative study on linear and region based diagrams. J. Spat. Inf. Sci. 2015; (10): 3–20.
- Lapovok I.A., Saleeva D.V., Kirichenko A.A., Murzakova A.V., Lopatukhin A.E., Kireev D.E. The study of dual HIV infection prevalence in Russia. Infektsionnye bolezni. 2020; 18(4): 138–48. https://doi.org/10.20953/1729-9225-2020-4-138-148 (in Russian)
- Los Alamos National Laboratory. RIP: Recombinant Identification Program. Available at: https://www.hiv.lanl.gov/content/sequence/RIP/RIP.html
- Pasechnik O.A., Blokh A.I. The prevalence of HIV recombinant forms in Russia and countries of the CIS: systematic review and metaanalysis. Infektsiya i immunitet. 2018; 8(2): 127–38. https://doi.org/10.15789/2220-7619-2018-2-127-138 (in Russian)
- Federal AIDS Center. Russian database. HIVDR in naive patients; 2020. Available at: http://www.hivrussia.info/wp-content/uploads/2020/12/2020-Rossijskaya-baza-dannyh-LU-VICH-u-naivnyh-patsientov.pdf (in Russian)
- Ostankova Yu.V., Shchemelev A.N., Zueva E.B., Churina M.A., Valutite D.E., Semenov A.V. HIV molecular epidemiology and pharmaco-resistance in patients with antiretroviral therapy failure in Arkhangelsk district. VICh infektsiya i immunosupressii. 2019; 11(4): 65–72. https://doi.org/10.22328/2077-9828-2019-11-4-79-90 (in Russian)
- Shchemelev A.N., Semenov A.V., Ostankova Yu.V., Zueva E.B., Valutite D.E., Semenova D.A., et al. Genetic diversity and drug resistance mutations of HIV-1 in Leningrad region. Zhurnal mikrobiologii, epidemiologii i immunobiologii. 2022; 99(1): 28–37. https://doi.org/10.36233/0372-9311-216 (in Russian)
- Hung M., Tokarsky E.J., Lagpacan L., Zhang L., Suo Z., Lansdon E.B. Elucidating molecular interactions of L-nucleotides with HIV-1 reverse transcriptase and mechanism of M184V-caused drug resistance. Commun. Biol. 2019; 2: 469. https://doi.org/10.1038/s42003-019-0706-x
- De Luca A., Giambenedetto S.D., Trotta M.P., Colafigli M., Prosperi M., Ruiz L., et al. Improved interpretation of genotypic changes in the HIV-1 reverse transcriptase coding region that determine the virological response to didanosine. J. Infect. Dis. 2007; 196(11): 1645–53. https://doi.org/10.1086/522231
- Lanier E.R., Givens N., Stone C., Griffin P., Gibb D., Walker S., et al. Effect of concurrent zidovudine use on the resistance pathway selected by abacavir-containing regimens. HIV Med. 2004; 5(6): 394–9. https://doi.org/10.1111/j.1468-1293.2004.00243.x
- Hu Z., Giguel F., Hatano H., Reid P., Lu J., Kuritzkes D.R. Fitness comparison of thymidine analog resistance pathways in human immunodeficiency virus type 1. J. Virol. 2006; 80(14): 7020–7. https://doi.org/10.1128/jvi.02747-05
- Ibe S., Sugiura W. Clinical significance of HIV reverse-transcriptase inhibitor-resistance mutations. Future Microbiol. 2011; 6(3): 295–315. https://doi.org/10.2217/fmb.11.7
- Xu H.T., Colby-Germinario S.P., Huang W., Oliveira M., Han Y., Quan Y., et al. Role of the K101E substitution in HIV-1 reverse transcriptase in resistance to rilpivirine and other nonnucleoside reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 2013; 57(11): 5649–57. https://doi.org/10.1128/aac.01536-13
- Madruga J.V., Cahn P., Grinsztejn B., Haubrich R., Lalezari J., Mills A., et al. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-1: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet. 2007; 370(9581): 29–38. https://doi.org/10.1016/s0140-6736(07)61047-2
- Archer R.H., Wisniewski M., Bambara R.A., Demeter L.M. The Y181C mutant of HIV-1 reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors alters the size distribution of RNase H cleavages. Biochemistry. 2001; 40(13): 4087–95. https://doi.org/10.1021/bi002328a
- Kolomeets A.N., Varghese V., Lemey P., Bobkova M.R., Shafer R.W. A uniquely prevalent nonnucleoside reverse transcriptase inhibitor resistance mutation in Russian subtype A HIV-1 viruses. AIDS. 2014; 28(17): F1–8. https://doi.org/10.1097/qad.0000000000000485
- Ostankova Yu.V., Shchemelev A.N., Zueva E.B., Churina M.A., Valutite D.E., Semenov A.V. HIV molecular epidemiology and pharmaco-resistance in patients with antiretroviral therapy failure in Arkhangelsk district. VICh infektsiya i immunosupressii. 2019; 11(4): 65–72. https://doi.org/10.22328/2077-9828-2019-11-4-79-9 (in Russian)
- Churina M.A., Ostankova Yu.V., Semenov A.V., Nikitina N.A., Rosolovskiy A.P., Grebenkina E.V., et al. HIV-1 drug-resistance and molecular epidemiology in patients with art failure in Veliky Novgorod. VICh-infektsiya i immunosupressii. 2017; 9(1): 82–92. https://doi.org/10.22328/2077-9828-2017-9-1-82-92 (in Russian)
- Chen M., Zhu Q., Xing H., Chen H., Jin X., Dong L., et al. The characteristics of pretreatment HIV-1 drug resistance in western Yunnan, China. Epidemiol. Infect. 2020; 148: e102. https://doi.org/10.1017/s095026882000093x
- Cheung K.W., Peng Q., He L., Cai K., Jiang Q., Zhou B., et al. Rapid and simultaneous detection of major drug resistance mutations in reverse transcriptase gene for HIV-1 CRF01_AE, CRF07_BC and subtype B in China using sequenom MassARRAY® system. PLoS One. 2016; 11(4): e0153641. https://doi.org/10.1371/journal.pone.0153641
Supplementary files